

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0234.13
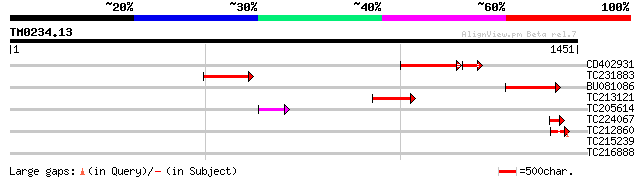
(1451 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (ja... 162 4e-44
TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment... 140 5e-33
BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza s... 134 2e-31
TC213121 93 1e-18
TC205614 59 2e-08
TC224067 44 5e-04
TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and... 44 7e-04
TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%) 35 0.18
TC216888 similar to UP|Q6YV21 (Q6YV21) Uridine kinase-like prote... 35 0.31
>CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 617
Score = 162 bits (410), Expect(2) = 4e-44
Identities = 89/156 (57%), Positives = 113/156 (72%)
Frame = +2
Query: 1001 GTGKTFLWKTLSYRLRSERKIVLNVASSGITSLLLPGGRTAHSLFSIPLVLNEDSCCNIR 1060
GT KTFLWKTLS +RS + ++ + + SLLLP G TAHS FSIPLV+ +DS CNI
Sbjct: 11 GTSKTFLWKTLSAGMRS-KGLMSSSCLKRLASLLLPRG*TAHSTFSIPLVIKDDSTCNIN 187
Query: 1061 LGSNKAELLKHTSLIIWDEAPMVNRWAFEAVDRTLRDIMEVGDVYGGGKPFGGKVVVLGG 1120
GS +++LL HT LIIWDE P++N++ FEA+DRTL+DIM + KPFGGK VVLGG
Sbjct: 188 QGSARSKLLLHTKLIIWDETPVMNKFCFEALDRTLQDIMASQNKDNATKPFGGK-VVLGG 364
Query: 1121 DFRQTLPIIPKASREEIVMATINSSRLWKFCKVLKL 1156
DFRQ LP+I K SR++IV + IN+S+ KVLKL
Sbjct: 365 DFRQILPVIRKGSRQDIVGSAINASK-----KVLKL 457
Score = 36.2 bits (82), Expect(2) = 4e-44
Identities = 20/51 (39%), Positives = 31/51 (60%)
Frame = +1
Query: 1158 ENMCLHGNDSLHDCEKLVEFSKWILDIGDGNLGDYNDGECDLDIPHDLMVP 1208
+NM L ++ D + + EF WIL I DGN + ++GE +DIP +L+ P
Sbjct: 460 KNMRLGTTNNNEDRDDIREFVDWILMIRDGNRDEEDEGE--IDIPKNLLNP 606
>TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment), partial
(17%)
Length = 1263
Score = 140 bits (352), Expect = 5e-33
Identities = 63/128 (49%), Positives = 90/128 (70%)
Frame = +1
Query: 495 MYTIEFQKRGLPHAHILIWLAPGSKLTTPEKIDSVICAELPDPVASPKLFEVVSMFMVHG 554
M+TIEFQKRGLPH +L++L P ++ + ++ID +I AE+P P+L+ +V MVHG
Sbjct: 877 MHTIEFQKRGLPHIQLLLFLHPDNQYPSSDEIDHIISAEIPSHEDDPELYTLVQNHMVHG 1056
Query: 555 PCGSSRKNSPCMVNGRCSKFFPKKYVDKTSFDIDGYPTYRRRNTGVFVERRDVQLDNGYV 614
PCG + +SPCM G+CS+F+PK ++ T D++GY YRRRN G + + V +DN Y+
Sbjct: 1057 PCGILQSHSPCMKEGKCSRFYPKIFLPNTLLDLNGYLVYRRRNDGRTILKNSVIVDNRYL 1236
Query: 615 VPYNAKLL 622
VPYNAKLL
Sbjct: 1237 VPYNAKLL 1260
>BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 426
Score = 134 bits (338), Expect = 2e-31
Identities = 67/141 (47%), Positives = 94/141 (66%)
Frame = +2
Query: 1269 SDSVVKVDEDVAIDANWITIEFLNGIKGSGLPDHRLCLKIGTPIMLMRNIDVSAGLCNGT 1328
SDS+ K D + + IT EF+N + S LP+H++ LK+ + IML+RN+D + GLCN T
Sbjct: 2 SDSIEKSDTIESEGFSTITTEFINSLSTSRLPNHKIKLKVDSRIMLLRNLDQNEGLCNST 181
Query: 1329 RLIVLDLRPNLIYGKVLNGNKAGTKTYIPRMTIVPSDSGLHVKVQRRQFPVCVCFAMTIN 1388
RLI+ ++I K+++G G YIPR+ PS S K+ RR+FP+ V +AMTIN
Sbjct: 182 RLIITMFADHIIEAKIMSGKGQGNTVYIPRLATSPSQSPWPFKLIRRKFPIIVSYAMTIN 361
Query: 1389 KSQGQTLSRVGVFLPRPVFSH 1409
KSQGQ L+ VG++LP PVFSH
Sbjct: 362 KSQGQLLASVGLYLPTPVFSH 424
>TC213121
Length = 713
Score = 92.8 bits (229), Expect = 1e-18
Identities = 45/110 (40%), Positives = 69/110 (61%)
Frame = +2
Query: 928 KDFPGIPYPISDEVPQFENVMLFNELRFDIDDMSVKHNDHLMKLNNGQRKVYDEVIDAVN 987
++FP + YPI V + N +++NEL +D D ++ + L + ++++++ +
Sbjct: 281 RNFPLMSYPIGYIVNPYGNKLIYNELAYDKDILAA*FSRCYHSLTDEHASIFNKIMHMIA 460
Query: 988 KSDGGFYFVYGSGGTGKTFLWKTLSYRLRSERKIVLNVASSGITSLLLPG 1037
GG YF+YG GGTGKTF+WKTLS + S IVL +ASSGI S+LLPG
Sbjct: 461 TQSGGVYFLYGYGGTGKTFVWKTLSSTIHSNSGIVLTMASSGILSMLLPG 610
Score = 41.6 bits (96), Expect = 0.003
Identities = 24/77 (31%), Positives = 40/77 (51%)
Frame = +3
Query: 680 CRYLSACEAAWRSFSFRIHDHWPPVQRLPFHMPNKQVVLYGNEEPIDRVVQRGQMSETMF 739
C Y+S E + +F +H P V+ L FH+ N+ + I ++ + + E++F
Sbjct: 30 CIYVSPPETC*KISAFPMHGRAPTVECLYFHLENQ*P--*KESQQIGSMLSKSTIKESIF 203
Query: 740 TGWMVANMIYEHGRHLT 756
T M +N +Y HGR LT
Sbjct: 204 TACMDSNKMYHHGRDLT 254
>TC205614
Length = 742
Score = 58.5 bits (140), Expect = 2e-08
Identities = 32/81 (39%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Frame = +2
Query: 637 SNCIKYLFKYINKGVDRV-TVSMKNECNEGQNVPEVDEIQQYY-DCRYLSACEAAWRSFS 694
SN ++F YI+ + S+K + +V YY DC+Y+S CEA WR FS
Sbjct: 497 SNLFSFIFHYISMVENHN*RTSLKLKDLASIEASQVSFPHMYYLDCQYISPCEACWRIFS 676
Query: 695 FRIHDHWPPVQRLPFHMPNKQ 715
F+I+ P V+RL FH+PN+Q
Sbjct: 677 FQIYGRIPIVERLYFHLPNEQ 739
>TC224067
Length = 415
Score = 43.9 bits (102), Expect = 5e-04
Identities = 27/41 (65%), Positives = 31/41 (74%), Gaps = 2/41 (4%)
Frame = -2
Query: 1381 VCFAMTINKSQGQTLSRV-GVFLPRPV-FSHGQLYVALSRV 1419
VC+ M INKS GQTLS+V GV LPRP SHGQ YV L++V
Sbjct: 360 VCYTMKINKSHGQTLSQVGGVLLPRPN**SHGQ-YVTLNQV 241
>TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (8%)
Length = 596
Score = 43.5 bits (101), Expect = 7e-04
Identities = 29/57 (50%), Positives = 35/57 (60%), Gaps = 8/57 (14%)
Frame = +2
Query: 1383 FAMTINKSQGQTLSRVGVFLPRPVFSHGQLYVALSRVKSRDGL--------KIYVDQ 1431
+AM+I+K QG TL RV L R F G +YVALSRV+S +GL KI VDQ
Sbjct: 5 WAMSIHKCQGMTLERVHTDLSR-AFGCGMVYVALSRVRSLEGLHLSAFNRSKIKVDQ 172
>TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%)
Length = 340
Score = 35.4 bits (80), Expect = 0.18
Identities = 16/22 (72%), Positives = 18/22 (81%)
Frame = -3
Query: 1373 QRRQFPVCVCFAMTINKSQGQT 1394
QR FP+ VCFAMT NKS+GQT
Sbjct: 338 QR**FPLIVCFAMTTNKSEGQT 273
>TC216888 similar to UP|Q6YV21 (Q6YV21) Uridine kinase-like protein, partial
(42%)
Length = 1425
Score = 34.7 bits (78), Expect = 0.31
Identities = 26/81 (32%), Positives = 35/81 (43%), Gaps = 7/81 (8%)
Frame = +1
Query: 66 TLLWDLIMENDVRSREFLANIRSYNSAFAFTSFGGKIESGLN------DGGGPPQFVISG 119
T L D I + R EF + I NS FA + + L +G GP F ++G
Sbjct: 670 TQLSDQISTLNERMDEFTSRIEELNSKFAISKDSAASQQNLALQAEACNGSGPTSFFVTG 849
Query: 120 -QNYHRIGSLLPNVGETPKFA 139
N GSLLPN + + A
Sbjct: 850 LSNGSLTGSLLPNSSSSSQLA 912
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,605,695
Number of Sequences: 63676
Number of extensions: 1012774
Number of successful extensions: 4728
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 4677
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4723
length of query: 1451
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1342
effective length of database: 5,698,948
effective search space: 7647988216
effective search space used: 7647988216
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0234.13


