

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0233.16
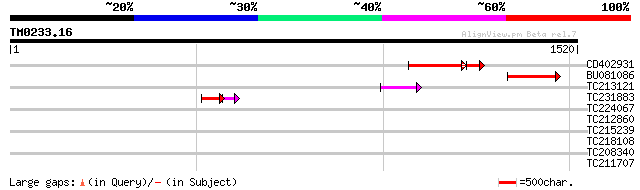
(1520 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (ja... 160 6e-45
BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza s... 138 2e-32
TC213121 95 3e-19
TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment... 68 9e-12
TC224067 40 0.006
TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and... 39 0.013
TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%) 32 2.8
TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete 30 6.2
TC208340 UP|Q9FRU5 (Q9FRU5) Nitrate transporter NRT1-2, partial ... 30 8.0
TC211707 30 8.0
>CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 617
Score = 160 bits (406), Expect(2) = 6e-45
Identities = 90/156 (57%), Positives = 113/156 (71%)
Frame = +2
Query: 1068 GTGKTFLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIP 1127
GT KTFLWKTL+ +RS K ++ + +ASLLLP G TAHS FSIPL + +DS C I
Sbjct: 11 GTSKTFLWKTLSAGMRS-KGLMSSSCLKRLASLLLPRG*TAHSTFSIPLVIKDDSTCNIN 187
Query: 1128 QGSPKAKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGG 1187
QGS ++KLL LIIWDE P+++++ FEALDRTL+DIM + +++ KPFGGK VVLGG
Sbjct: 188 QGSARSKLLLHTKLIIWDETPVMNKFCFEALDRTLQDIMASQNKDNATKPFGGK-VVLGG 364
Query: 1188 DFRQILPVIPKGRRAEIVMSTINSSRLWRFCKVLNL 1223
DFRQILPVI KG R +IV S IN+S+ KVL L
Sbjct: 365 DFRQILPVIRKGSRQDIVGSAINASK-----KVLKL 457
Score = 40.4 bits (93), Expect(2) = 6e-45
Identities = 18/49 (36%), Positives = 32/49 (64%)
Frame = +1
Query: 1225 QNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNVQIPKDLL 1273
+NMRL T+ + D ++++ F W+L + DG + ++GE + IPK+LL
Sbjct: 460 KNMRLGTTNNNEDRDDIREFVDWILMIRDGNRDEEDEGE--IDIPKNLL 600
>BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 426
Score = 138 bits (347), Expect = 2e-32
Identities = 71/141 (50%), Positives = 92/141 (64%)
Frame = +2
Query: 1336 SDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMRNLDISTGLCNGT 1395
SDS+ + D +TTEF+N L S +P+HK+ LK + IML+RNLD + GLCN T
Sbjct: 2 SDSIEKSDTIESEGFSTITTEFINSLSTSRLPNHKIKLKVDSRIMLLRNLDQNEGLCNST 181
Query: 1396 RLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQFPLIASFAMTIN 1455
RLI+ ++I A ++SG G VYI R+ PS P K RR+FP+I S+AMTIN
Sbjct: 182 RLIITMFADHIIEAKIMSGKGQGNTVYIPRLATSPSQSPWPFKLIRRKFPIIVSYAMTIN 361
Query: 1456 KSQGQSLSQVGVYLPKPVFSH 1476
KSQGQ L+ VG+YLP PVFSH
Sbjct: 362 KSQGQLLASVGLYLPTPVFSH 424
>TC213121
Length = 713
Score = 94.7 bits (234), Expect = 3e-19
Identities = 47/112 (41%), Positives = 68/112 (59%)
Frame = +2
Query: 993 SLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVEMSNLHDECFSKLNDGQLKIYQEIITA 1052
S ++F M + + YGN L+ NEL +D ++ C+ L D I+ +I+
Sbjct: 275 SHRNFPLMSYPIGYIVNPYGNKLIYNELAYDKDILAA*FSRCYHSLTDEHASIFNKIMHM 454
Query: 1053 INANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQIILNVASSGIASLLLPG 1104
I +GG +F+YGYGGTGKTF+WKTL+ + S I+L +ASSGI S+LLPG
Sbjct: 455 IATQSGGVYFLYGYGGTGKTFVWKTLSSTIHSNSGIVLTMASSGILSMLLPG 610
>TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment), partial
(17%)
Length = 1263
Score = 67.8 bits (164), Expect(2) = 9e-12
Identities = 32/60 (53%), Positives = 41/60 (68%)
Frame = +1
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
+TIEFQKRGLPH +LL+L +N+ S ID +I AE+P PEL+ V N+MVHGP
Sbjct: 880 HTIEFQKRGLPHIQLLLFLHPDNQYPSSDEIDHIISAEIPSHEDDPELYTLVQNHMVHGP 1059
Score = 21.9 bits (45), Expect(2) = 9e-12
Identities = 13/44 (29%), Positives = 19/44 (42%)
Frame = +3
Query: 572 HGPWSMWIE**RFSLHEEWSVLQILPQKICFKNNI***WLSDIS 615
+G W MW FS++E V L I + + WL +S
Sbjct: 1044 YGTWPMWNSTISFSMYERRQVQSFLS*NISAQYSFRFKWLFGLS 1175
>TC224067
Length = 415
Score = 40.4 bits (93), Expect = 0.006
Identities = 36/87 (41%), Positives = 47/87 (53%), Gaps = 4/87 (4%)
Frame = -2
Query: 1428 LMPSDESMPVKFQRRQFPLIASFAMTINKSQGQSLSQV-GVYLPKPV-FSHGQLYVAISR 1485
L+ D KFQ + + M INKS GQ+LSQV GV LP+P SHGQ YV +++
Sbjct: 414 LISFDRLASFKFQ--DVNSLVCYTMKINKSHGQTLSQVGGVLLPRPN**SHGQ-YVTLNQ 244
Query: 1486 VKS--RAGLKILICNEDTSQLDVTKNI 1510
V S R K + C + Q +KNI
Sbjct: 243 V*SY*RRKSKSINCEKYDLQRIFSKNI 163
>TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (8%)
Length = 596
Score = 39.3 bits (90), Expect = 0.013
Identities = 22/57 (38%), Positives = 35/57 (60%)
Frame = +2
Query: 1449 SFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTSQLD 1505
++AM+I+K QG +L +V L + F G +YVA+SRV+S GL + N ++D
Sbjct: 2 AWAMSIHKCQGMTLERVHTDLSR-AFGCGMVYVALSRVRSLEGLHLSAFNRSKIKVD 169
>TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%)
Length = 340
Score = 31.6 bits (70), Expect = 2.8
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = -3
Query: 1440 QRRQFPLIASFAMTINKSQGQS 1461
QR FPLI FAMT NKS+GQ+
Sbjct: 338 QR**FPLIVCFAMTTNKSEGQT 273
>TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete
Length = 1122
Score = 30.4 bits (67), Expect = 6.2
Identities = 11/38 (28%), Positives = 23/38 (59%)
Frame = -2
Query: 1231 TSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNVQI 1268
+STC + E+ ++ W LD G+ G ++G+S +++
Sbjct: 488 SSTCKYAS*ELSYYISWYLDQGESSGGGKSNGDSRIEM 375
>TC208340 UP|Q9FRU5 (Q9FRU5) Nitrate transporter NRT1-2, partial (98%)
Length = 2068
Score = 30.0 bits (66), Expect = 8.0
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 4/39 (10%)
Frame = +3
Query: 1038 LNDGQLKIYQEIITAINANNGGFFFV----YGYGGTGKT 1072
LN G+L + IITA+ N G+F + Y Y GTG +
Sbjct: 1593 LNKGRLDYFYYIITALEVVNFGYFILCAKWYKYKGTGSS 1709
>TC211707
Length = 631
Score = 30.0 bits (66), Expect = 8.0
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Frame = -3
Query: 1135 LLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQI-- 1192
++ +L++ P+ S+ A + R++ K HNS K V++L GDF+QI
Sbjct: 212 IMNFLALLVLQSVPLRSQGAVRGYPK--RNLGEIK-HNSPHKIILANVLILDGDFKQIGL 42
Query: 1193 LPVIPKGR 1200
PV+ +G+
Sbjct: 41 QPVLGEGK 18
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.359 0.162 0.586
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,724,814
Number of Sequences: 63676
Number of extensions: 1116389
Number of successful extensions: 12896
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 7008
Number of HSP's successfully gapped in prelim test: 598
Number of HSP's that attempted gapping in prelim test: 5580
Number of HSP's gapped (non-prelim): 8233
length of query: 1520
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1410
effective length of database: 5,635,272
effective search space: 7945733520
effective search space used: 7945733520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0233.16


