

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
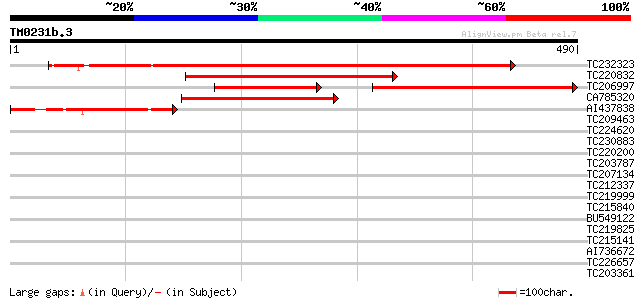
Query= TM0231b.3
(490 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232323 UP|PUR1_SOYBN (P52418) Amidophosphoribosyltransferase, ... 652 0.0
TC220832 similar to UP|PUR1_SOYBN (P52418) Amidophosphoribosyltr... 339 2e-93
TC206997 similar to UP|Q39000 (Q39000) Amidophosphoribosyltransf... 327 6e-90
CA785320 253 2e-67
AI437838 homologue to PIR|E84541|E845 amidophosphoribosyltransfe... 134 1e-31
TC209463 35 0.058
TC224620 similar to UP|O23254 (O23254) Serine hydroxymethyltrans... 35 0.058
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 34 0.17
TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 34 0.17
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 33 0.22
TC207134 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 33 0.29
TC212337 weakly similar to UP|Q9SM91 (Q9SM91) Zwh18.1, partial (... 33 0.38
TC219999 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (5%) 32 0.49
TC215840 homologue to UP|Q41122 (Q41122) Proline-rich protein pr... 32 0.49
BU549122 32 0.49
TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process... 32 0.49
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 32 0.64
AI736672 weakly similar to GP|21284389|gb| acyl-CoA binding prot... 32 0.84
TC226657 similar to UP|O81098 (O81098) RNA polymerase I, II and ... 32 0.84
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 32 0.84
>TC232323 UP|PUR1_SOYBN (P52418) Amidophosphoribosyltransferase, chloroplast
precursor (Glutamine phosphoribosylpyrophosphate
amidotransferase) (ATASE) (GPAT) , partial (72%)
Length = 1231
Score = 652 bits (1683), Expect = 0.0
Identities = 338/407 (83%), Positives = 361/407 (88%), Gaps = 3/407 (0%)
Frame = +3
Query: 34 LHNPTSFPNPTTLSHAHKPPYPSPC--KNHNTTFPEPLCNDDEDQKPREECGVVGIYGDP 91
LHN S PN +LSH P P C KN NT + DDEDQKPREECGVVGIYGDP
Sbjct: 27 LHNK-SLPNQNSLSHKLSSPLPLACNPKNQNTC----VFFDDEDQKPREECGVVGIYGDP 191
Query: 92 EASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIG 151
EASRLC LALHALQHRGQEGAGIV VH+N + QSVTGVGLV+DVF ++KL +LPGT AIG
Sbjct: 192 EASRLCSLALHALQHRGQEGAGIVAVHDNHL-QSVTGVGLVSDVFEQSKLSRLPGTSAIG 368
Query: 152 HVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLE-ENGSIFNTSSDTE 210
HVRYSTAGQSMLKNVQPF+A YRF +V VAHNGN VNYRSLR +LE NGSIFNT+SDTE
Sbjct: 369 HVRYSTAGQSMLKNVQPFLADYRFAAVAVAHNGNFVNYRSLRARLEHNNGSIFNTTSDTE 548
Query: 211 VVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGA 270
VVLHLIATSK RPF+LRIVDACE L+GAYS VFVTEDKLVAVRDPFGFRPLVMG+R+NGA
Sbjct: 549 VVLHLIATSKHRPFLLRIVDACEHLQGAYSLVFVTEDKLVAVRDPFGFRPLVMGRRTNGA 728
Query: 271 VVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSS 330
VV ASETCALDLIEA YEREVYPGEV+VVD G+QSLCLVSHPEPKQCIFEHIYF+LP+S
Sbjct: 729 VVLASETCALDLIEATYEREVYPGEVIVVDHTGIQSLCLVSHPEPKQCIFEHIYFALPNS 908
Query: 331 VVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHY 390
VVFG+SVYESR+ FGEILA+ESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHY
Sbjct: 909 VVFGRSVYESRKKFGEILASESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHY 1088
Query: 391 VGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSS 437
VGRTFIEPSQKIRD GVKLKLSPV AVLEGKRVVVVDDSIVRGTTSS
Sbjct: 1089VGRTFIEPSQKIRDFGVKLKLSPVHAVLEGKRVVVVDDSIVRGTTSS 1229
>TC220832 similar to UP|PUR1_SOYBN (P52418) Amidophosphoribosyltransferase,
chloroplast precursor (Glutamine
phosphoribosylpyrophosphate amidotransferase) (ATASE)
(GPAT) , partial (32%)
Length = 551
Score = 339 bits (869), Expect = 2e-93
Identities = 165/183 (90%), Positives = 178/183 (97%)
Frame = +2
Query: 153 VRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVV 212
VRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR+LR LE++GSIFNT+SDTEVV
Sbjct: 2 VRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRTLRTNLEDSGSIFNTTSDTEVV 181
Query: 213 LHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVV 272
LHLIATSK RPFILRIVDACEKL+GAYS VFVTEDKLVAVRDPFGFRPLVMG+RSNG+VV
Sbjct: 182 LHLIATSKHRPFILRIVDACEKLEGAYSIVFVTEDKLVAVRDPFGFRPLVMGRRSNGSVV 361
Query: 273 FASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVV 332
FASETCALDLIEA YEREVYPGEVLVVDKNG+QSLCL+SHP+PKQCIFEHIYF+LP+SVV
Sbjct: 362 FASETCALDLIEATYEREVYPGEVLVVDKNGIQSLCLMSHPQPKQCIFEHIYFALPNSVV 541
Query: 333 FGK 335
FG+
Sbjct: 542 FGR 550
>TC206997 similar to UP|Q39000 (Q39000) Amidophosphoribosyltransferase
(Fragment), partial (61%)
Length = 1313
Score = 327 bits (839), Expect = 6e-90
Identities = 165/177 (93%), Positives = 173/177 (97%)
Frame = +3
Query: 314 EPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGY 373
+PKQCI EHIYF+LP+SVVFG+SVYESRR FGEILATESPVECDVVIAVPDSGVVAALGY
Sbjct: 312 QPKQCISEHIYFALPNSVVFGRSVYESRRQFGEILATESPVECDVVIAVPDSGVVAALGY 491
Query: 374 AAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRG 433
AAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKLSPV+AVLEGKRVVVVDDSIVRG
Sbjct: 492 AAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVRAVLEGKRVVVVDDSIVRG 671
Query: 434 TTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
TTSSKIVRL+KEAGAKEVHMRIA PPIIGSCYYGVDTPSSEELISNRMSV+EIR+FI
Sbjct: 672 TTSSKIVRLLKEAGAKEVHMRIASPPIIGSCYYGVDTPSSEELISNRMSVEEIRDFI 842
Score = 154 bits (388), Expect = 1e-37
Identities = 77/92 (83%), Positives = 81/92 (87%)
Frame = +1
Query: 178 VGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKG 237
V VAHNGNLV Y+SL LE+N SIF T+SDTEVVLHL TSKQRPF+LRIVDACEKLKG
Sbjct: 7 VRVAHNGNLVKYKSL*TNLEDNVSIFTTTSDTEVVLHLTGTSKQRPFLLRIVDACEKLKG 186
Query: 238 AYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNG 269
AYSFVFVTEDKLV VRDPFGFRPLV GKRSNG
Sbjct: 187 AYSFVFVTEDKLVVVRDPFGFRPLVTGKRSNG 282
>CA785320
Length = 409
Score = 253 bits (645), Expect = 2e-67
Identities = 125/136 (91%), Positives = 132/136 (96%)
Frame = +2
Query: 149 AIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSD 208
+IGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNY++LR LE+NGSIFNT+SD
Sbjct: 2 SIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYKTLRTNLEDNGSIFNTTSD 181
Query: 209 TEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSN 268
TEVVLHLIATSK RPFILRIVDACEKL+GAYS VFVTEDKLVAVRDPFGFRPLVMG+RSN
Sbjct: 182 TEVVLHLIATSKHRPFILRIVDACEKLEGAYSIVFVTEDKLVAVRDPFGFRPLVMGRRSN 361
Query: 269 GAVVFASETCALDLIE 284
GAVVFASETCALDLIE
Sbjct: 362 GAVVFASETCALDLIE 409
>AI437838 homologue to PIR|E84541|E845 amidophosphoribosyltransferase
[imported] - Arabidopsis thaliana, partial (13%)
Length = 434
Score = 134 bits (336), Expect = 1e-31
Identities = 82/150 (54%), Positives = 97/150 (64%), Gaps = 5/150 (3%)
Frame = +3
Query: 1 MAASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKN 60
MAA+ +T SSS S S F +T P + P P A KPP KN
Sbjct: 21 MAAAAAATTLSSSLSKPSSLTF---------QTHSFPITLPTPPRA--AAKPPLLVFSKN 167
Query: 61 H-----NTTFPEPLCNDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIV 115
++TF +D+KPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIV
Sbjct: 168 PISDIVSSTFSRAEEGAWDDEKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIV 347
Query: 116 TVHNNKIFQSVTGVGLVADVFTEAKLDQLP 145
TV+NN + QS+TGVGLV++VF ++KLDQLP
Sbjct: 348 TVNNN-VLQSITGVGLVSEVFNQSKLDQLP 434
>TC209463
Length = 434
Score = 35.4 bits (80), Expect = 0.058
Identities = 19/46 (41%), Positives = 27/46 (58%)
Frame = +3
Query: 11 SSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPS 56
SS+SS + S P +TP N+T H TSFP P ++ + PP P+
Sbjct: 186 SSASSHALSSPSLSTP----NRTHHPSTSFPPPLSILSSTSPPPPA 311
>TC224620 similar to UP|O23254 (O23254) Serine hydroxymethyltransferase
(Serine methylase) (Glycine hydroxymethyltransferase)
(SHMT) , complete
Length = 1887
Score = 35.4 bits (80), Expect = 0.058
Identities = 24/54 (44%), Positives = 31/54 (56%)
Frame = +1
Query: 2 AASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYP 55
AA N ST S +S++ + SKP T+TP PG T +PT P PT+ PP P
Sbjct: 292 AAMNTSTRSKTSAAHAPSKPSTSTP-NPGAST-SSPTPAPRPTS------PPTP 429
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 33.9 bits (76), Expect = 0.17
Identities = 20/47 (42%), Positives = 24/47 (50%), Gaps = 2/47 (4%)
Frame = +2
Query: 19 SKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPS--PCKNHNT 63
SKP + P N PTS P P T S + PPYPS PC+ +T
Sbjct: 317 SKPSASAPTLLSNPGPSPPTSLP-PLT*STSTSPPYPSPVPCRTFST 454
>TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (40%)
Length = 506
Score = 33.9 bits (76), Expect = 0.17
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 1/74 (1%)
Frame = +2
Query: 1 MAASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFP-NPTTLSHAHKPPYPSPCK 59
+AA+ + S + + + P +++P FP + + +FP +PT H PP PSP
Sbjct: 182 IAATAATAAESPAPTPTPDFPPSSSPPFPSPASNPSTANFPPSPTPAPSHHSPPAPSPSS 361
Query: 60 NHNTTFPEPLCNDD 73
N + + P P DD
Sbjct: 362 NPSPS-PAPAPVDD 400
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 33.5 bits (75), Expect = 0.22
Identities = 23/67 (34%), Positives = 33/67 (48%)
Frame = +3
Query: 2 AASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNH 61
A+S+ S+ SSSSSS S S P +++ P + P S L PYP P +
Sbjct: 372 ASSSSSSSSSSSSSSS*SSPSSSSSSSPFSSAPPAPPSGAFFELLLLLELLPYPPPSSDD 551
Query: 62 NTTFPEP 68
N++ P P
Sbjct: 552 NSSSPSP 572
>TC207134 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} ,
partial (67%)
Length = 1418
Score = 33.1 bits (74), Expect = 0.29
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 1/65 (1%)
Frame = +2
Query: 2 AASNFSTLSSSSSSFSKSKPF-TNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKN 60
A +N S ++ SSS SKP T C P + T SFP+PT+ + HK +PC+
Sbjct: 26 APANTSWRNTESSS---SKPT*TTRRCSPSSST-----SFPSPTSSTSQHKRASATPCRT 181
Query: 61 HNTTF 65
+ T+
Sbjct: 182PSPTW 196
>TC212337 weakly similar to UP|Q9SM91 (Q9SM91) Zwh18.1, partial (11%)
Length = 536
Score = 32.7 bits (73), Expect = 0.38
Identities = 19/52 (36%), Positives = 24/52 (45%)
Frame = +3
Query: 7 STLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPC 58
S+LSSS+ + KP N P GN+ +S P P PP P PC
Sbjct: 213 SSLSSSAWAPPSPKPTKNAP---GNRQTLKSSSLPPPPPPPQTPNPPSPPPC 359
>TC219999 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (5%)
Length = 641
Score = 32.3 bits (72), Expect = 0.49
Identities = 17/47 (36%), Positives = 25/47 (53%)
Frame = -3
Query: 11 SSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSP 57
S+ ++ S P +TP +L PT P+PTT S ++ PP P P
Sbjct: 360 STQTAASVPIPEPSTPSVQTFLSLRPPTPSPSPTTSSPSYPPPPPPP 220
>TC215840 homologue to UP|Q41122 (Q41122) Proline-rich protein precursor,
partial (74%)
Length = 1171
Score = 32.3 bits (72), Expect = 0.49
Identities = 17/57 (29%), Positives = 24/57 (41%)
Frame = +2
Query: 1 MAASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSP 57
+A SN + + S P + P LH P + P+P H H PP P+P
Sbjct: 98 VAVSNVFAVELELETLPPSYPHPHPVHSPTPAPLHPPANAPHPHHHHHHHHPPAPAP 268
>BU549122
Length = 670
Score = 32.3 bits (72), Expect = 0.49
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Frame = +3
Query: 5 NFSTLSSSSSSFSKSKPFTNT----PCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKN 60
NFS + + + S PF++T P +K+L N FPNP+ S+ S K
Sbjct: 222 NFSNFHRTIGTTTSSTPFSSTPHPSPLTTSSKSLSNYLPFPNPSNFSNT------SQQKP 383
Query: 61 HNT 63
HNT
Sbjct: 384 HNT 392
>TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process protein
MID2 (Serine-rich protein SMS1) (Protein kinase A
interference protein), partial (19%)
Length = 862
Score = 32.3 bits (72), Expect = 0.49
Identities = 23/64 (35%), Positives = 32/64 (49%), Gaps = 3/64 (4%)
Frame = +2
Query: 4 SNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPT---SFPNPTTLSHAHKPPYPSPCKN 60
S+ ST SSSS S S P ++P P +P+ S P+P + S + P Y SP
Sbjct: 35 SSSSTSSSSSPSPSSPPPPPSSPSPPSTPPRPSPSPPPSPPSPASSSASSSPSYGSPSS* 214
Query: 61 HNTT 64
+TT
Sbjct: 215SSTT 226
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 32.0 bits (71), Expect = 0.64
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 7/50 (14%)
Frame = +2
Query: 24 NTPCFPGNKTLHNP-------TSFPNPTTLSHAHKPPYPSPCKNHNTTFP 66
NT C T P TS P+PT+ + H+P +PC+ N T P
Sbjct: 461 NTKCSSWKVTSTTPPCSRSSSTSSPSPTSSTWPHRPASATPCRTPNPTSP 610
>AI736672 weakly similar to GP|21284389|gb| acyl-CoA binding protein
{Arabidopsis thaliana}, partial (11%)
Length = 419
Score = 31.6 bits (70), Expect = 0.84
Identities = 19/54 (35%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Frame = +2
Query: 14 SSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNHNTT-FP 66
S S F++ P P+SFP+ TT+S + PP P P + TT FP
Sbjct: 152 SGSPSSNRFSSASFSPTFSPSSFPSSFPSKTTISPSRAPPPPKPPQPRATTPFP 313
>TC226657 similar to UP|O81098 (O81098) RNA polymerase I, II and III 24.3 kDa
subunit (AT3g22320/MCB17_5) , complete
Length = 773
Score = 31.6 bits (70), Expect = 0.84
Identities = 21/51 (41%), Positives = 26/51 (50%)
Frame = +1
Query: 7 STLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSP 57
ST SS +S S S+P TP KT P+S P PT+ PP P+P
Sbjct: 238 STSSSPTSPRSASRP*RPTPTA*TPKTSIEPSSSPRPTS------PPSPAP 372
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 31.6 bits (70), Expect = 0.84
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Frame = +3
Query: 2 AASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHA--------HKPP 53
++S+ S SSSSSS S S +++ P + + +P S P S A P
Sbjct: 315 SSSSSSPPSSSSSSSSSSSSSSSS*SSPSSSSSSSPFSSAPPAPPSGAFFELLLLLELLP 494
Query: 54 YPSPCKNHNTTFPEP 68
YP P + N++ P P
Sbjct: 495 YPPPSSDDNSSSPSP 539
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,872,838
Number of Sequences: 63676
Number of extensions: 373448
Number of successful extensions: 3449
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 3069
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3362
length of query: 490
length of database: 12,639,632
effective HSP length: 101
effective length of query: 389
effective length of database: 6,208,356
effective search space: 2415050484
effective search space used: 2415050484
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0231b.3


