

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.5
(406 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
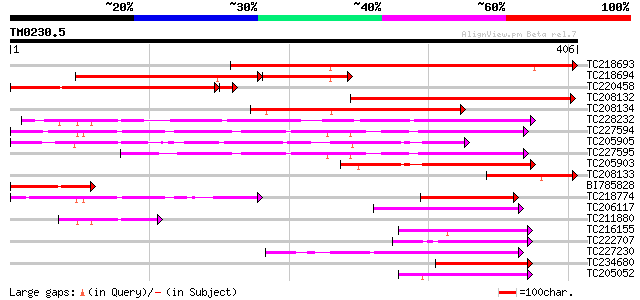
Score E
Sequences producing significant alignments: (bits) Value
TC218693 similar to UP|Q94KA8 (Q94KA8) BZIP transcription factor... 387 e-108
TC218694 homologue to UP|Q94KA8 (Q94KA8) BZIP transcription fact... 245 6e-85
TC220458 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription fact... 278 3e-77
TC208132 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription fact... 263 8e-71
TC208134 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription fact... 261 3e-70
TC228232 homologue to UP|Q43449 (Q43449) G-box binding factor, c... 197 7e-51
TC227594 UP|Q43450 (Q43450) G-box binding factor (Fragment), com... 185 3e-47
TC205905 similar to UP|Q41113 (Q41113) BZIP transcriptional repr... 165 4e-41
TC227595 homologue to UP|Q43451 (Q43451) G-box binding factor (F... 119 2e-27
TC205903 similar to UP|Q41113 (Q41113) BZIP transcriptional repr... 118 5e-27
TC208133 similar to UP|Q94KA7 (Q94KA7) BZIP transcription factor... 107 1e-23
BI785828 100 9e-22
TC218774 similar to UP|Q43450 (Q43450) G-box binding factor (Fra... 90 2e-18
TC206117 weakly similar to UP|Q9FUD3 (Q9FUD3) BZIP protein BZO2H... 65 6e-11
TC211880 homologue to UP|Q43449 (Q43449) G-box binding factor, p... 64 9e-11
TC216155 weakly similar to UP|Q8L5W2 (Q8L5W2) BZIP transcription... 63 2e-10
TC222707 homologue to UP|Q9AT29 (Q9AT29) BZip transcription fact... 62 4e-10
TC227230 similar to UP|P93839 (P93839) G/HBF-1, partial (72%) 61 8e-10
TC234680 similar to UP|Q9AT29 (Q9AT29) BZip transcription factor... 61 8e-10
TC205052 similar to UP|Q8L5W2 (Q8L5W2) BZIP transcription factor... 60 2e-09
>TC218693 similar to UP|Q94KA8 (Q94KA8) BZIP transcription factor 2, partial
(61%)
Length = 1226
Score = 387 bits (993), Expect = e-108
Identities = 199/257 (77%), Positives = 218/257 (84%), Gaps = 9/257 (3%)
Frame = +3
Query: 159 EHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVH 218
E GKTPG SANG HSKSGES S+GTSEGSD NS NDS+LKSG R+DSFEDEPSQNG++VH
Sbjct: 3 EPGKTPGESANGIHSKSGESASDGTSEGSDENSHNDSQLKSGERQDSFEDEPSQNGSSVH 182
Query: 219 TTQNGGLNTP----HQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGN 274
QNG N +QTMS++PIS+ A GAVPGP TNLNIGMDYWGTPTSS IPALHG
Sbjct: 183 APQNGVHNRSQTVVNQTMSILPISSTSARGAVPGPTTNLNIGMDYWGTPTSSTIPALHGK 362
Query: 275 VPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQ 334
VPSTAVAGGM GSRDGVQSQ WLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQ
Sbjct: 363 VPSTAVAGGMIAAGSRDGVQSQVWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQ 542
Query: 335 RAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL-----PGNDDLRSGKSDQHT 389
RAEALKEENA+LRSEVS+IRS+YEQL +ENAALKERLG++ PG +DLRSG++DQH
Sbjct: 543 RAEALKEENATLRSEVSQIRSEYEQLRSENAALKERLGDIPGVATPGKEDLRSGQNDQHV 722
Query: 390 HDDAQQSGQTEAVQGGH 406
+D QQSGQTE VQG H
Sbjct: 723 SNDTQQSGQTEVVQGVH 773
>TC218694 homologue to UP|Q94KA8 (Q94KA8) BZIP transcription factor 2,
partial (46%)
Length = 653
Score = 245 bits (626), Expect(2) = 6e-85
Identities = 116/137 (84%), Positives = 122/137 (88%), Gaps = 3/137 (2%)
Frame = +3
Query: 48 MPPHGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFA 107
+PPHGFLAS+PQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFA
Sbjct: 6 IPPHGFLASSPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFA 185
Query: 108 MPSPNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSL--GSLNM-ITRKNNEHGKTP 164
M SPNGI +ASGN PGS+E GKPPEVKEKLPIKRSKGS G+LNM IT KNNE GK P
Sbjct: 186 MASPNGIADASGNAPGSIEVGGKPPEVKEKLPIKRSKGSASGGNLNMWITGKNNEPGKIP 365
Query: 165 GTSANGAHSKSGESGSE 181
G SANG HSKSGES S+
Sbjct: 366 GESANGIHSKSGESASD 416
Score = 87.4 bits (215), Expect(2) = 6e-85
Identities = 45/68 (66%), Positives = 52/68 (76%), Gaps = 4/68 (5%)
Frame = +1
Query: 182 GTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQTMSMVPI 237
GTSEGSD NSQNDS+LKS R+DSFEDEPSQNG++VH QNG N P +QTMS++PI
Sbjct: 418 GTSEGSDENSQNDSQLKSRERQDSFEDEPSQNGSSVHAPQNGVHNRPQTVVNQTMSILPI 597
Query: 238 SAGGAPGA 245
APGA
Sbjct: 598 XTTXAPGA 621
>TC220458 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription factor 3,
partial (41%)
Length = 776
Score = 278 bits (710), Expect(2) = 3e-77
Identities = 131/151 (86%), Positives = 136/151 (89%), Gaps = 1/151 (0%)
Frame = +2
Query: 1 MGSSEMDKTPKEKESKTPPL-TPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQ 59
MGSSEMDKT KEKESKTPP T QEQ TT TGT+NP +W FQAYSP+PPHGFLASNPQ
Sbjct: 287 MGSSEMDKTTKEKESKTPPPPTSQEQSSTTGTGTINP-EWPGFQAYSPIPPHGFLASNPQ 463
Query: 60 AHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASG 119
AHPYMWGVQ MPPYGTPPHPYVAMYP GGIYAHPS+PPGSYPF+PFAMPSPNGI EASG
Sbjct: 464 AHPYMWGVQQFMPPYGTPPHPYVAMYPPGGIYAHPSMPPGSYPFNPFAMPSPNGIAEASG 643
Query: 120 NTPGSMEADGKPPEVKEKLPIKRSKGSLGSL 150
NTPGSMEADGKPPEVKEKLPIKRSKGSLG L
Sbjct: 644 NTPGSMEADGKPPEVKEKLPIKRSKGSLGRL 736
Score = 29.3 bits (64), Expect(2) = 3e-77
Identities = 12/13 (92%), Positives = 12/13 (92%)
Frame = +3
Query: 151 NMITRKNNEHGKT 163
NMIT KNNEHGKT
Sbjct: 738 NMITGKNNEHGKT 776
>TC208132 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription factor 3,
partial (33%)
Length = 706
Score = 263 bits (673), Expect = 8e-71
Identities = 137/162 (84%), Positives = 143/162 (87%), Gaps = 1/162 (0%)
Frame = +1
Query: 245 AVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERE 304
AVPGP TNLNIGMDYWGTP SSNIP L VPSTAVAGGM T GSRD QSQ WLQDERE
Sbjct: 1 AVPGPTTNLNIGMDYWGTPGSSNIPGLGRKVPSTAVAGGMVTVGSRDSAQSQLWLQDERE 180
Query: 305 LKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTEN 364
LKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV+RIRSDYEQLL+EN
Sbjct: 181 LKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVNRIRSDYEQLLSEN 360
Query: 365 AALKERLGELPGNDD-LRSGKSDQHTHDDAQQSGQTEAVQGG 405
AALKERLGELP NDD RS ++DQH +D QQSGQTEA+ GG
Sbjct: 361 AALKERLGELPPNDDHHRSCRNDQHVGNDTQQSGQTEAMPGG 486
>TC208134 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription factor 3,
partial (39%)
Length = 570
Score = 261 bits (668), Expect = 3e-70
Identities = 134/160 (83%), Positives = 140/160 (86%), Gaps = 6/160 (3%)
Frame = +3
Query: 173 SKSGESGSEG--TSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPH- 229
SKSGES SEG TSEGSDANSQNDS+LKSGGR+DSFEDEPSQNG+ +T QNGGLNTPH
Sbjct: 3 SKSGESASEGEGTSEGSDANSQNDSQLKSGGRQDSFEDEPSQNGSLAYTAQNGGLNTPHT 182
Query: 230 ---QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMAT 286
QTMS++PISAGGAPGAVPGP TNLNIGMDYWGTP SSNIPAL VPSTAVAGGM T
Sbjct: 183 VVNQTMSIIPISAGGAPGAVPGPTTNLNIGMDYWGTPASSNIPALGRKVPSTAVAGGMVT 362
Query: 287 GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQ 326
GSRD QSQ WLQDERELKRQRRKQSNRESARRSRLRKQ
Sbjct: 363 VGSRDSAQSQLWLQDERELKRQRRKQSNRESARRSRLRKQ 482
>TC228232 homologue to UP|Q43449 (Q43449) G-box binding factor, complete
Length = 1723
Score = 197 bits (501), Expect = 7e-51
Identities = 141/381 (37%), Positives = 192/381 (50%), Gaps = 13/381 (3%)
Frame = +1
Query: 9 TPKEKESKTPPLTPQEQPPTTSTGTV----NPPDWSS-FQAYSP--MPPHGFLASN---P 58
T +E +K P +P +TS+ + + PDWSS QAY P F ASN P
Sbjct: 43 TGEESTAKVP------KPSSTSSIQIPLAPSYPDWSSSMQAYYAPGATPPAFFASNIASP 204
Query: 59 QAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEA 117
H YMWG QH ++PPY TP PY A+YP G +YAHPS+
Sbjct: 205 TPHSYMWGSQHPLIPPYSTPV-PYPAIYPPGNVYAHPSM------------------AMT 327
Query: 118 SGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGE 177
T E GK + K+++ K SK + K ++GK N S+S E
Sbjct: 328 LSTTQNGTEFVGKGSDEKDRVSAKSSKAVSANNGS---KAGDNGKASSGPRNDGTSQSAE 498
Query: 178 SGSEGTSEGSDANS-QNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVP 236
SGSEG+S+ SD N+ Q +S G D + G N + ++S++P
Sbjct: 499 SGSEGSSDASDENTNQQESATNKKGSFDQMLVD--------------GANARNNSVSIIP 636
Query: 237 ISAGGAPGAVPGPATNLNIGMDYWG-TPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQS 295
A P T+LNIGM+ W +P + N S AV G R+
Sbjct: 637 QPGNPAVSMSP---TSLNIGMNLWNASPAGDEAAKMRQNQSSGAVTPPTIMG--REVALG 801
Query: 296 QHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRS 355
+HW+QDERELK+Q+RKQSNRESARRSRLRKQAEC+EL +R E+L EN +LR E+ R+
Sbjct: 802 EHWIQDERELKKQKRKQSNRESARRSRLRKQAECEELQKRVESLGSENQTLREELQRVSE 981
Query: 356 DYEQLLTENAALKERLGELPG 376
+ ++L +EN ++KE L L G
Sbjct: 982 ECKKLTSENDSIKEELERLCG 1044
>TC227594 UP|Q43450 (Q43450) G-box binding factor (Fragment), complete
Length = 1872
Score = 185 bits (470), Expect = 3e-47
Identities = 137/385 (35%), Positives = 195/385 (50%), Gaps = 14/385 (3%)
Frame = +2
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSP---MPPH--GFLA 55
MG+SE +K+ K + P T ++ T PDW++ Q Y P +PP+ +A
Sbjct: 245 MGNSEEEKSVKTGSPSSSPATTEQ---TNQPNIHVYPDWAAMQYYGPRVNIPPYFNSAVA 415
Query: 56 SNPQAHPYMWGV-QHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
S HPYMWG Q +M PYG P Y A Y HGG+Y HP++ G + SP
Sbjct: 416 SGHAPHPYMWGPPQPMMQPYGPP---YAAFYSHGGVYTHPAVAIGPHSHGQGVPSSP--- 577
Query: 115 VEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
+ TP S+E+ K + +K+ KG I N E ++ GA ++
Sbjct: 578 ---AAGTPSSVESPTKFSGNTNQGLVKKLKGFDELAMSIGNCNAE------SAERGAENR 730
Query: 175 SGES-GSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLN----TPH 229
+S +EG+S+GSD N+ ++ K R+ S E P + QNG + +
Sbjct: 731 LSQSVDTEGSSDGSDGNTAGANQTK---RKRSREGTPITDAEGKTELQNGPASKETASSK 901
Query: 230 QTMSMVPISAGGA---PGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMAT 286
+ +S P S G P G AT L + N +H ST+ A A
Sbjct: 902 KIVSATPASVAGTLVGPVVSSGMATALEL----------RNPSTVHSKANSTSAAQPCAV 1051
Query: 287 GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASL 346
V+++ WLQ+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ E L EN SL
Sbjct: 1052------VRNETWLQNERELKRERRKQSNRESARRSRLRKQAETEELARKVEMLTAENVSL 1213
Query: 347 RSEVSRIRSDYEQLLTENAALKERL 371
+SE++R+ EQ+ EN+AL+E+L
Sbjct: 1214KSEITRLTEGSEQMRMENSALREKL 1288
>TC205905 similar to UP|Q41113 (Q41113) BZIP transcriptional repressor ROM1,
partial (83%)
Length = 1226
Score = 165 bits (417), Expect = 4e-41
Identities = 132/338 (39%), Positives = 158/338 (46%), Gaps = 9/338 (2%)
Frame = +1
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSS-FQAY-----SPMPPHGFL 54
MG+ E K S TP + PDWSS QAY +P P
Sbjct: 379 MGAGEESTAKSSKVSSAAQETPT---------ALAYPDWSSSMQAYYAPGGTPPPFFAST 531
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
++P HPY+WG QH +MPPYGTP PY A+YP G IYAHPS+ PS
Sbjct: 532 VASPTPHPYLWGSQHPLMPPYGTPV-PYPAIYPPGSIYAHPSMAVN---------PS--- 672
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
IV+ + E +GK E K + K+ KG + K E GK S N S
Sbjct: 673 IVQQN------TEIEGKGAEGKYRDSSKKLKGPSANT---ASKAGESGKAGSGSGNDGIS 825
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
+SGESGSEG+S SD N+ + ++ SF+ A Q S
Sbjct: 826 QSGESGSEGSSNASDENTNQQES--AANKKGSFDLMLVDGANA-------------QNNS 960
Query: 234 MVPISAGGAPG--AVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRD 291
IS PG VP PATNLNIGMD W + A + S A G+A G
Sbjct: 961 AGAISQSSVPGKPVVPMPATNLNIGMDLWNASSGGAEAAKMRHNQSGAP--GVALG---- 1122
Query: 292 GVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAEC 329
W+QDERELKRQ RKQSNRES RRSRLRKQAEC
Sbjct: 1123----DQWVQDERELKRQIRKQSNRESPRRSRLRKQAEC 1224
>TC227595 homologue to UP|Q43451 (Q43451) G-box binding factor (Fragment),
complete
Length = 1453
Score = 119 bits (298), Expect = 2e-27
Identities = 100/300 (33%), Positives = 143/300 (47%), Gaps = 8/300 (2%)
Frame = +3
Query: 80 PYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADGKPPEVKEKLP 139
P + +YP P P SP A TP S+E+ K +
Sbjct: 81 PNIHVYPDWAAMQGPHSHGQGVPSSPAA------------GTPSSVESPTKFSGNTNQGL 224
Query: 140 IKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGES-GSEGTSEGSDANSQNDSELK 198
+K+ KG I N E ++ GA ++ +S +EG+S+GSD N+ ++ K
Sbjct: 225 VKKLKGFDELAMSIGNCNAE------SAERGAENRLSQSVDTEGSSDGSDGNTAGANQTK 386
Query: 199 SGGRRDSFEDEPSQNGTAVHTTQNGGLN----TPHQTMSMVPISAGGA---PGAVPGPAT 251
R+ S E P+ + QNG + + + +S P S G P G AT
Sbjct: 387 ---RKRSREGTPTTDAEGKTELQNGPASKETSSSKKIVSATPASVAGTLVGPVVSSGMAT 557
Query: 252 NLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRK 311
L + N +H ST+ A V ++ LQ+ERELKR+RRK
Sbjct: 558 TLEL----------RNPSTVHSKANSTSAPQPCAI------VPNETCLQNERELKRERRK 689
Query: 312 QSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERL 371
QSNRESARRSRLRKQAE +ELA++ E L EN SL+SE++R+ EQ+ EN+AL+E+L
Sbjct: 690 QSNRESARRSRLRKQAETEELARKVEMLTAENVSLKSEITRLTEGSEQMRMENSALREKL 869
>TC205903 similar to UP|Q41113 (Q41113) BZIP transcriptional repressor ROM1,
partial (46%)
Length = 827
Score = 118 bits (295), Expect = 5e-27
Identities = 73/146 (50%), Positives = 91/146 (62%), Gaps = 7/146 (4%)
Frame = +1
Query: 238 SAGG-APGAVPG------PATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
SAG + +VPG PATNLNIGMD W + A + S A G+A G
Sbjct: 37 SAGAISQXSVPGKPVXXMPATNLNIGMDLWNASSGGAEAAKMRHNQSGAP--GVALG--- 201
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
W+QDERELKRQ+RKQSNRESARRSRLRKQAEC+EL +R E+L EN +LR E+
Sbjct: 202 -----DQWVQDERELKRQKRKQSNRESARRSRLRKQAECEELQKRVESLGGENQTLREEL 366
Query: 351 SRIRSDYEQLLTENAALKERLGELPG 376
R+ + E+L +EN ++KE L L G
Sbjct: 367 QRLSEECEKLTSENNSIKEELERLCG 444
>TC208133 similar to UP|Q94KA7 (Q94KA7) BZIP transcription factor 3, partial
(14%)
Length = 386
Score = 107 bits (266), Expect = 1e-23
Identities = 55/67 (82%), Positives = 60/67 (89%), Gaps = 2/67 (2%)
Frame = +2
Query: 342 ENASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDD--LRSGKSDQHTHDDAQQSGQT 399
ENASLRSEV+RIRSDYEQLL+ENAALKERLGELP NDD RSG++DQH +D QQSGQT
Sbjct: 2 ENASLRSEVNRIRSDYEQLLSENAALKERLGELPPNDDHHHRSGRNDQHVGNDTQQSGQT 181
Query: 400 EAVQGGH 406
EAVQGGH
Sbjct: 182 EAVQGGH 202
>BI785828
Length = 428
Score = 100 bits (250), Expect = 9e-22
Identities = 47/61 (77%), Positives = 52/61 (85%)
Frame = +2
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQA 60
MGSS+MDKTPKEKESKTPP T QEQ TT+ T N PDWS+FQ YSP+PPHGFLAS+PQA
Sbjct: 248 MGSSDMDKTPKEKESKTPPATSQEQSSTTAMPTTN-PDWSNFQTYSPIPPHGFLASSPQA 424
Query: 61 H 61
H
Sbjct: 425 H 427
>TC218774 similar to UP|Q43450 (Q43450) G-box binding factor (Fragment),
partial (85%)
Length = 1334
Score = 89.7 bits (221), Expect = 2e-18
Identities = 65/188 (34%), Positives = 88/188 (46%), Gaps = 7/188 (3%)
Frame = +1
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS----PMPPH--GFL 54
MG+SE +K+ K E + P+T + T T PDW++ QAY MPP+ +
Sbjct: 163 MGNSEEEKSTKT-EKPSSPVTVDQANQTNQTNIHVYPDWAAMQAYYGPRVTMPPYYNSAV 339
Query: 55 ASNPQAHPYMWGV-QHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
AS HPYMWG Q +MPPYG P Y A+YPHGG+Y HP++P G S SP
Sbjct: 340 ASGHAPHPYMWGPPQPMMPPYGPP---YAAIYPHGGVYTHPAVPIGPLTHSQGVPSSP-- 504
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
+ TP S+E PP+ S G+ + K + S H+
Sbjct: 505 ----AAGTPLSIET---PPK------------SSGNTDQGLMKKLKEFDGLAMSIGNGHA 627
Query: 174 KSGESGSE 181
+S E G E
Sbjct: 628 ESAERGGE 651
Score = 89.4 bits (220), Expect = 3e-18
Identities = 42/70 (60%), Positives = 58/70 (82%)
Frame = +2
Query: 295 SQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIR 354
++ W+Q+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ E+L ENA+L+SE++R+
Sbjct: 878 AEAWVQNERELKRERRKQSNRESARRSRLRKQAETEELARKVESLNAENATLKSEINRLT 1057
Query: 355 SDYEQLLTEN 364
E++ N
Sbjct: 1058ESSEKMRVGN 1087
>TC206117 weakly similar to UP|Q9FUD3 (Q9FUD3) BZIP protein BZO2H2, partial
(48%)
Length = 1225
Score = 65.1 bits (157), Expect = 6e-11
Identities = 39/108 (36%), Positives = 55/108 (50%)
Frame = +3
Query: 261 GTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARR 320
G+P S+N P N A +G D + + ++KR RRK SNR+SARR
Sbjct: 285 GSPVSANKPEGRENHTKGATSGSSEPSDEDDEAGACEQSTNPADMKRLRRKVSNRDSARR 464
Query: 321 SRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALK 368
SR RKQA+ +L + E LK ENA+L + + + + T N LK
Sbjct: 465 SRRRKQAQLSDLELQVEKLKVENATLYKQFTDASQHFREADTNNRVLK 608
>TC211880 homologue to UP|Q43449 (Q43449) G-box binding factor, partial (24%)
Length = 726
Score = 64.3 bits (155), Expect = 9e-11
Identities = 37/80 (46%), Positives = 45/80 (56%), Gaps = 6/80 (7%)
Frame = +3
Query: 36 PPDWSSFQAYSP--MPPHGFLASN---PQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGG 89
P SS QAY P F ASN P H YMWG QH ++PPY TP PY A+YP G
Sbjct: 483 PSSTSSIQAYYAPGATPPAFFASNIASPTPHSYMWGSQHPLIPPYSTPV-PYPAIYPPGN 659
Query: 90 IYAHPSIPPGSYPFSPFAMP 109
+YAHPS+ S+ + +P
Sbjct: 660 VYAHPSMAMVSHLLTLIPLP 719
>TC216155 weakly similar to UP|Q8L5W2 (Q8L5W2) BZIP transcription factor
ATB2, partial (66%)
Length = 1264
Score = 63.2 bits (152), Expect = 2e-10
Identities = 36/99 (36%), Positives = 56/99 (56%), Gaps = 3/99 (3%)
Frame = +1
Query: 279 AVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQ---SNRESARRSRLRKQAECDELAQR 335
A GG T S + +R++ QR+++ SNRESARRSR+RKQ + L+ +
Sbjct: 586 ASPGGSGTYSSGSSSLQNSGSEGDRDIMEQRKRKRMLSNRESARRSRIRKQQHLEGLSAQ 765
Query: 336 AEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
+ LK+ENA + + +S Y + ENA L+ ++GEL
Sbjct: 766 LDQLKKENAQINTNISITTQMYLNVEAENAILRAQMGEL 882
>TC222707 homologue to UP|Q9AT29 (Q9AT29) BZip transcription factor, complete
Length = 671
Score = 62.4 bits (150), Expect = 4e-10
Identities = 38/100 (38%), Positives = 56/100 (56%)
Frame = +3
Query: 275 VPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQ 334
VP ++ +T D +Q + DER + RR SNRESARRSR+RKQ DEL
Sbjct: 240 VPPSSCLSSNSTSDEADEIQFN--IIDER---KHRRMISNRESARRSRMRKQKHLDELWS 404
Query: 335 RAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
+ L+ EN +L +++ + ++++L ENA LKE L
Sbjct: 405 QVVRLRTENHNLIDKLNHVSESHDRVLQENARLKEEASAL 524
>TC227230 similar to UP|P93839 (P93839) G/HBF-1, partial (72%)
Length = 1250
Score = 61.2 bits (147), Expect = 8e-10
Identities = 51/185 (27%), Positives = 80/185 (42%)
Frame = +2
Query: 184 SEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVPISAGGAP 243
+ GS A SQ+ S GG + + N + V + QT S I + P
Sbjct: 194 TRGSLAKSQDPSPFSEGGSQPT-------NPSLVES----------QTTSKGSIPSENDP 322
Query: 244 GAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDER 303
+ TN+ +G+ P PA+ ++ + + +G S + D
Sbjct: 323 SKLQDKDTNVPVGIP--SIPAMQKKPAVAIRPSTSGSSREQSDDEDIEGETSMNDNTDPA 496
Query: 304 ELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTE 363
++KR RR SNRESARRSR RKQA +L + L+ EN++L ++ + Y +
Sbjct: 497 DVKRVRRMLSNRESARRSRRRKQAHLTDLETQVSQLRGENSTLLKRLTDVSQKYSDSAVD 676
Query: 364 NAALK 368
N LK
Sbjct: 677 NRVLK 691
>TC234680 similar to UP|Q9AT29 (Q9AT29) BZip transcription factor, partial
(49%)
Length = 571
Score = 61.2 bits (147), Expect = 8e-10
Identities = 31/69 (44%), Positives = 45/69 (64%)
Frame = +2
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENA 365
++ RR SNRESARRSR+RKQ DEL + L+ EN L +++ + ++Q++ ENA
Sbjct: 347 RKHRRMISNRESARRSRMRKQKHLDELWSQVVWLRNENHQLMDKLNHVSESHDQVMQENA 526
Query: 366 ALKERLGEL 374
LKE+ EL
Sbjct: 527 QLKEQALEL 553
>TC205052 similar to UP|Q8L5W2 (Q8L5W2) BZIP transcription factor ATB2,
partial (58%)
Length = 953
Score = 60.1 bits (144), Expect = 2e-09
Identities = 35/100 (35%), Positives = 57/100 (57%), Gaps = 4/100 (4%)
Frame = +1
Query: 279 AVAGGMATGGSRDGVQ----SQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQ 334
A + G ++G + +Q S+ LQ E ++++R SNRESARRSR+RKQ D+LA
Sbjct: 508 ACSSGTSSGATSSMLQNNSGSEEELQALMEQRKRKRMISNRESARRSRMRKQKHLDDLAS 687
Query: 335 RAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
+ L+ EN + + V+ Y + EN+ L+ ++ EL
Sbjct: 688 QVTQLRNENHQILTSVNLTTQKYLAVEAENSVLRAQVNEL 807
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,794,065
Number of Sequences: 63676
Number of extensions: 347982
Number of successful extensions: 2995
Number of sequences better than 10.0: 396
Number of HSP's better than 10.0 without gapping: 2328
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2665
length of query: 406
length of database: 12,639,632
effective HSP length: 99
effective length of query: 307
effective length of database: 6,335,708
effective search space: 1945062356
effective search space used: 1945062356
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0230.5


