

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0228a.4
(1904 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
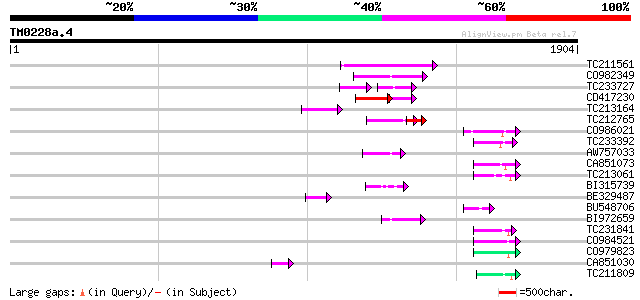
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 166 1e-40
CO982349 119 2e-26
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 71 3e-25
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 104 4e-22
TC213164 92 2e-18
TC212765 91 5e-18
CO986021 75 4e-13
TC233392 72 2e-12
AW757033 72 2e-12
CA851073 69 3e-11
TC213061 67 7e-11
BI315739 66 1e-10
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 65 3e-10
BU548706 64 5e-10
BI972659 62 2e-09
TC231841 60 1e-08
CO984521 58 3e-08
CO979823 57 8e-08
CA851030 56 1e-07
TC211809 56 1e-07
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase (Fragment)
, partial (59%)
Length = 1077
Score = 166 bits (419), Expect = 1e-40
Identities = 103/327 (31%), Positives = 172/327 (52%)
Frame = +1
Query: 1110 ILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVR 1169
++A EV+ +++ + K+D EKAYD V+W+FL ++ F P+ + I V+
Sbjct: 13 LIANEVID--EAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEECVK 186
Query: 1170 TPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSK 1229
+ ++SVL NGS F QRGLRQGDPL+P+LF + E L ++ VEE ++P V
Sbjct: 187 SASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLVGA 366
Query: 1230 DGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRR 1289
+GV IS L +ADD + F +A+K+ + IK L F S +++N KS V Q +
Sbjct: 367 NGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGVTDQWK 546
Query: 1290 QRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVC 1349
Q E + + S + YLG+P+ + + +II K +L+ WK ++ GRV
Sbjct: 547 Q-EAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGRVT 723
Query: 1350 LAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKG 1409
L KS+L+S+P++ +P SV + + K+ R +WG G + + + W+ + K +G
Sbjct: 724 LIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKERG 903
Query: 1410 RLGLRSARKNNEALLGKLVHSFLNEKG 1436
+ R +N + F++ G
Sbjct: 904 GI*KRLPNNHNAVANHNSIFCFIDFTG 984
>CO982349
Length = 795
Score = 119 bits (297), Expect = 2e-26
Identities = 83/255 (32%), Positives = 127/255 (49%), Gaps = 6/255 (2%)
Frame = +3
Query: 1154 FGFPPRTV-----DLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMER 1208
F FPP T I + + +VS+L N S + F QRGLRQGDPL+P LF + E
Sbjct: 27 FYFPP*TAIRH*QHWIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEG 206
Query: 1209 LAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEAS 1268
L ++ V+ ++ V K+ +S L +ADD + F +A+ + +++IK L F AS
Sbjct: 207 LTGLMREAVDRKRFNSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELAS 386
Query: 1269 GMKVNLEKSRMLFSKV-VGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATII 1327
G+K+N +SR F + +E + + S S YLG+P+ R+ II
Sbjct: 387 GLKINFAESR--FGAIWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPII 560
Query: 1328 EKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGK 1387
K T+LA WK ++ GRV L +IL++LP++ P V + + R +WG
Sbjct: 561 RKCETKLARWKQRHISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGG 740
Query: 1388 GGNARSWHLVAWKDV 1402
G R V W+ V
Sbjct: 741 GSXQRRIAWVKWETV 785
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 70.9 bits (172), Expect(2) = 3e-25
Identities = 39/137 (28%), Positives = 73/137 (52%), Gaps = 5/137 (3%)
Frame = -2
Query: 1235 SHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELS 1294
SH+ ADD+++FC+ + + + + + ++ G ++ KS++ + GQR R +S
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRL-RVIS 275
Query: 1295 GLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSI 1354
L+ + Y G+P+ +G+ R H + +K+ +LASWK ++L+ GR+ L S+
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 1355 -----LSSLPVHAMQSL 1366
L S V+A Q L
Sbjct: 94 IHGMLLYSFSVYAWQVL 44
Score = 64.7 bits (156), Expect(2) = 3e-25
Identities = 40/109 (36%), Positives = 64/109 (58%), Gaps = 1/109 (0%)
Frame = -3
Query: 1106 RDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIM 1165
+D + EV++ + +K GG +ALKID+ KA+D ++ DFL + L FG+ + I
Sbjct: 831 KDCTCVTSEVINMLD-KKVFGGNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIR 655
Query: 1166 WGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLF-VLCMERLAIQI 1213
+ + +S+ NG + F QRG+RQGDPLSP L+ C+E+ I +
Sbjct: 654 VILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLYRQRCLEQGTIYV 508
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like protein
{Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 104 bits (259), Expect = 4e-22
Identities = 54/126 (42%), Positives = 86/126 (67%), Gaps = 1/126 (0%)
Frame = -3
Query: 1161 VDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEG 1220
V L+ + + T +S+LWNG L F G+RQ DP++PYLFVLC+ERL+ I ++ +
Sbjct: 686 VQLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKK 507
Query: 1221 KWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSR-M 1279
R ++V++ G+ ISHL F DD++LF +A+ DQ+ +I TL F ++SG KVN++K++
Sbjct: 506 L*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKSF 327
Query: 1280 LFSKVV 1285
LF K++
Sbjct: 326 LFPKML 309
Score = 67.4 bits (163), Expect = 6e-11
Identities = 34/90 (37%), Positives = 52/90 (57%)
Frame = -2
Query: 1275 EKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRL 1334
+ + FSK V + ++S +G+ +LGKYLG+P+ F II+KVN R
Sbjct: 342 QNQKFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRF 163
Query: 1335 ASWKNNLLNKAGRVCLAKSILSSLPVHAMQ 1364
+ WK+NLL+ GR+ L K ++ +LP H MQ
Sbjct: 162 SRWKSNLLSMVGRLTLTK*VV*ALPSHIMQ 73
>TC213164
Length = 446
Score = 92.4 bits (228), Expect = 2e-18
Identities = 50/136 (36%), Positives = 75/136 (54%)
Frame = +3
Query: 981 DKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGAN 1040
D LVA +EVR+ + + K+PGPDG + F K++W IL DL + + NG
Sbjct: 12 DLLVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFL 191
Query: 1041 PELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFI 1100
A L LIPKV +P L + +PISL YK++ K+L R + +L ++ Q F+
Sbjct: 192 KRSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFM 371
Query: 1101 LGRGTRDNIILAQEVM 1116
GR T N+++A E+M
Sbjct: 372 EGRHTLHNVVIANEIM 419
>TC212765
Length = 637
Score = 90.9 bits (224), Expect = 5e-18
Identities = 64/175 (36%), Positives = 95/175 (53%), Gaps = 1/175 (0%)
Frame = +3
Query: 1199 PYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIK 1258
PYLFVLCMERLA+ I +L + WRP+ VS D + +L DD+ LF A D L
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNRLVPYLMLVDDIFLFINAIGD*AYLAS 194
Query: 1259 HTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRV 1318
TL +F + G+K+NL+KS M S + + +S + I A +LGKYL L + GR
Sbjct: 195 LTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHGRS 374
Query: 1319 KRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLW-LPDSV 1372
K F ++E+++ + + L K +V + SS+P ++ LW LP S+
Sbjct: 375 KIVDF-LLVERIHNPWPRGRGSFLTK--QVNIV*RSWSSMP--SLFILWRLPGSL 524
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/65 (44%), Positives = 41/65 (62%)
Frame = +2
Query: 1334 LASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARS 1393
LASWK LLN+A + CL +L + P++ M++ WLP S+ +DK R IW K G + S
Sbjct: 416 LASWKGKLLNQASKHCLT*LVLHAFPIYPMEASWLPQSIFQAIDKASRVTIWNKVGASLS 595
Query: 1394 WHLVA 1398
WH V+
Sbjct: 596 WHSVS 610
>CO986021
Length = 900
Score = 74.7 bits (182), Expect = 4e-13
Identities = 62/213 (29%), Positives = 94/213 (44%), Gaps = 23/213 (10%)
Frame = -1
Query: 1524 QGAWNFDRLY--TLLPQE------IREEISSVQIPSIQTGEDRILWREASDDIYSASSAY 1575
Q WN+D + L E ++IS++ I Q +D +LW+ IYS SAY
Sbjct: 900 QNTWNWDFKWRRNLFDHENEQAIAFMDDISAISIH--QQLQDSMLWKADPTGIYSTKSAY 727
Query: 1576 QFLNEDGLP--ETGFWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLA-TTASCIR 1632
+ L + P + +K++WK P + F W + RD L A R H+A C
Sbjct: 726 RLLLPNNRPGQHSSNFKILWKLKIPPRAELFSWRLFRDRLSIRANLLRRHVALQDIMCPL 547
Query: 1633 CGAVVEDAEHVLRRCPGSV--------WVRGQLGRLLPWISDSATFRHWLFGHLQNRDNT 1684
CG E+A H+ C ++ W R LG L +A F + G R++
Sbjct: 546 CGNHQEEAGHLFFHCRMTIGLWWESMNWTR-TLGAFLD--DPAAHFIQFSDGFGAQRNHN 376
Query: 1685 LFC----AMAWNLWKWRNSFVFEQVPWQPGDVM 1713
C A+ +W+ RNS +F+ P+QP VM
Sbjct: 375 RRCLWWIALTSTIWQHRNSLLFKGTPFQPPKVM 277
>TC233392
Length = 1145
Score = 72.4 bits (176), Expect = 2e-12
Identities = 50/162 (30%), Positives = 72/162 (43%), Gaps = 15/162 (9%)
Frame = -1
Query: 1557 DRILWREASDDIYSASSAYQFLNE--DGLPETGFWKLIWKASAPEKVRFFLWLVGRDALP 1614
D +W+ +++YS SAY+ L +G + G + +WK P K F W + RD LP
Sbjct: 656 DTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGALQDLWKLKIPAKASIFAWRLIRDRLP 477
Query: 1615 TNAKRHRHHLATTAS-CIRCGAVVEDAEHVLRRC--------PGSVWVRGQLGRLLPWIS 1665
T + HR + S C C EDA H+ C WVR LG + I+
Sbjct: 476 TKSNLHRRQVVLEDSLCPFCRIREEDASHIFLECNKIRPLWWESQTWVR-SLG--VSPIN 306
Query: 1666 DSATFRHWLFG----HLQNRDNTLFCAMAWNLWKWRNSFVFE 1703
F + G NR T + A+ W+LWK RN +F+
Sbjct: 305 KRQHFLQHVHGTPGSKRYNRWKTWWIALTWSLWKHRNQVIFQ 180
>AW757033
Length = 441
Score = 72.0 bits (175), Expect = 2e-12
Identities = 50/144 (34%), Positives = 76/144 (52%), Gaps = 1/144 (0%)
Frame = -2
Query: 1185 FPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVL 1244
F RGLRQGDPL+P LF + E L ++ V ++ V K +S L +ADD +
Sbjct: 431 FSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADDTI 252
Query: 1245 LFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKV-VGQRRQRELSGLVGISRAS 1303
F +A+ + +R+IK L F ASG+K+N KSR F + V ++E + + S S
Sbjct: 251 FFGEATMENVRVIKTILRGFELASGLKINFAKSR--FGAISVPD*WRKEAAEFMNCSLLS 78
Query: 1304 NLGKYLGLPLLQGRVKRDHFATII 1327
YLG+P+ +R+ + II
Sbjct: 77 LPFSYLGIPIGANPRRRETWDPII 6
>CA851073
Length = 633
Score = 68.6 bits (166), Expect = 3e-11
Identities = 46/173 (26%), Positives = 79/173 (45%), Gaps = 15/173 (8%)
Frame = +1
Query: 1556 EDRILWREASDDIYSASSAYQFLNEDG--LPETGFWKLIWKASAPEKVRFFLWLVGRDAL 1613
+D ++W+ +YS SAY+ L + E +K IWK P + F W + +D L
Sbjct: 25 QDTMVWKADPCGVYSTKSAYRILMTCNRHVSEANIFKTIWKLKIPPRAAVFSWRLIKDRL 204
Query: 1614 PTNAKRHRHHLATTAS-CIRCGAVVEDAEHVLRRCPGSVWVRGQLGRLLPW--------I 1664
PT R +++ + C CG E+A+H+ C RG + W +
Sbjct: 205 PTRHNLLRRNVSIQENECPLCGYEQEEADHLFFNCK---MTRGLWWESMRWNQIVGPLSV 375
Query: 1665 SDSATFRHWLFGHLQNRDNTLFC----AMAWNLWKWRNSFVFEQVPWQPGDVM 1713
S ++ F + G R++T +C A+ ++WK RN +F+ ++P VM
Sbjct: 376 SPASHFVQFCDGFGAGRNHTRWCGWWIALTSSIWKHRNLLIFQGNQFEPSKVM 534
>TC213061
Length = 823
Score = 67.0 bits (162), Expect = 7e-11
Identities = 52/177 (29%), Positives = 72/177 (40%), Gaps = 19/177 (10%)
Frame = +2
Query: 1556 EDRILWREASDDIYSASSAYQFLNEDGLPETGF---WKLIWKASAPEKVRFFLWLVGRDA 1612
ED+ +W S Y A SAY+ + E G+PE +K +WK P KV F W + +D
Sbjct: 125 EDQWVWAAESSGSYLARSAYRVIRE-GIPEEEQDREFKELWKLKVPMKVTMFAWRLLKDI 301
Query: 1613 LPT--NAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPG--SVWVRGQLGRLLPWISDSA 1668
LPT N +R R L C C ++ E A H+ C +W L W+
Sbjct: 302 LPTRDNLRRKRVELHEYV-CPLCRSMDESASHLFFHCSKILPIWWES-----LSWVKLVG 463
Query: 1669 TFRHWLFGHLQ------------NRDNTLFCAMAWNLWKWRNSFVFEQVPWQPGDVM 1713
F H H NR + A+ W +WK RN +F + VM
Sbjct: 464 AFPHHPRHHFHQHSHEVYQGLQGNRWKWWWLALTWTIWKHRNDIIFSNATFNAHKVM 634
>BI315739
Length = 442
Score = 66.2 bits (160), Expect = 1e-10
Identities = 49/144 (34%), Positives = 76/144 (52%)
Frame = +3
Query: 1196 PLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLR 1255
PLSPYLF +CME+LAI IQ+ V E W P+++S++G GIS ++ + ++ F +
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNGPGISPIYSSQIIVYFL----*KPL 215
Query: 1256 LIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQ 1315
++K + C V E SR +F K G +R + L L LL
Sbjct: 216 VLKLNW*EMC--CNSFV*PEVSRRVF-KNPGF*HKR-------MCLIEKLPGSLL**LLA 365
Query: 1316 GRVKRDHFATIIEKVNTRLASWKN 1339
GR K++ FA +++ +N RL WK+
Sbjct: 366 GRTKKEDFAHVVDMINNRLDGWKS 437
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 65.1 bits (157), Expect = 3e-10
Identities = 33/88 (37%), Positives = 47/88 (52%)
Frame = -2
Query: 992 VRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLI 1051
++SA+ S K+PGPDG + F K +W L D+ + NG A+ + LI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 1052 PKVDNPTKLKDLRPISLCNVSYKLITKV 1079
PKV +P L D RPISL YK++ K+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>BU548706
Length = 647
Score = 64.3 bits (155), Expect = 5e-10
Identities = 32/106 (30%), Positives = 56/106 (52%)
Frame = +2
Query: 1523 RQGAWNFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDG 1582
+ G+WN+D L +LP + I + +PSI+ G D ++ + +++ ++ +S Y+
Sbjct: 242 QDGSWNYDFLEEMLPSHVVCIICGMLLPSIENGNDSLI*KFSTNGNFNLASIYR------ 403
Query: 1583 LPETGFWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTA 1628
+K IW P++V+ FLW DALPTN H ++ T A
Sbjct: 404 --NXXLFKSIWDRPGPKRVKLFLWKSAHDALPTNYLXHH*NIXTNA 535
>BI972659
Length = 453
Score = 62.4 bits (150), Expect = 2e-09
Identities = 40/151 (26%), Positives = 74/151 (48%), Gaps = 4/151 (2%)
Frame = +1
Query: 1248 QASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQ--RELSGLVGISRASNL 1305
+AS D + ++K L F SG+++N KS+ +VG + + + L+ +
Sbjct: 7 EASWDNIIVLKSMLRGFEMVSGLRINYAKSQF---GIVGFQPNWAHDAAQLLNCRQLDIP 177
Query: 1306 GKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQS 1365
YLG+P+ R + +I K +L+ W L+ GRV L KS+L++LP++ +
Sbjct: 178 FHYLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSF 357
Query: 1366 LWLPDSVCNHVDKMVRNCIWG--KGGNARSW 1394
+P + + + + R +WG + N SW
Sbjct: 358 FKIPQRIVDKLVSLQRTFMWGGNQHHNRISW 450
>TC231841
Length = 791
Score = 59.7 bits (143), Expect = 1e-08
Identities = 44/163 (26%), Positives = 68/163 (40%), Gaps = 17/163 (10%)
Frame = +3
Query: 1557 DRILWREASDDIYSASSAYQFLNEDGLPET--GFWKLIWKASAPEKVRFFLWLVGRDALP 1614
D+ +W+ Y+A SAY L + E G ++ +WK P K+ F W + RD LP
Sbjct: 63 DQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDGVFEELWKLKLPSKITIFAWRLIRDRLP 242
Query: 1615 TNAKRHRHHL-ATTASCIRCGAVVEDAEHVLRRCP--GSVWVRGQLGRLLPWISDSATF- 1670
T + R + C C + E A H+ C VW L W++ F
Sbjct: 243 TRSNLRRKQIEVDDPRCPFCRSAEESAAHLFFHCSRIAPVWWES-----LSWVNLLGVFP 407
Query: 1671 ----RHWL-------FGHLQNRDNTLFCAMAWNLWKWRNSFVF 1702
+H+L G +R + A+ W +WK RN+ +F
Sbjct: 408 NHPRQHFLQHIYGVTAGMRASRWKWWWLALTWTIWKQRNNMIF 536
>CO984521
Length = 716
Score = 58.2 bits (139), Expect = 3e-08
Identities = 48/170 (28%), Positives = 74/170 (43%), Gaps = 13/170 (7%)
Frame = -2
Query: 1557 DRILWREASDDIYSASSAYQFLNED--GLPETGFWKLIWKASAPEKVRFFLWLVGRDALP 1614
D+ W YS SAY+ L+ G + G +K +WK P KV F W + +D LP
Sbjct: 646 DQWKWAAEPSGCYSTKSAYKALHHVTVGEEQDGKFKELWKLRVPLKVAIFAWRLIQDKLP 467
Query: 1615 TNAKRHRHHLATTA-SCIRCGAVVEDAEHVLRRCP--GSVWVRGQ----LGRLLPWISDS 1667
T A + + C C +V E A H+ C +W Q + + P+ D
Sbjct: 466 TKANLRKKRVELQEYLCPLCRSVEETASHLFFHCSKVSPLWWESQSWVNMMGVFPYQPDQ 287
Query: 1668 ATFRHWLFG---HLQ-NRDNTLFCAMAWNLWKWRNSFVFEQVPWQPGDVM 1713
+H +FG LQ R + A+ +++WK RNS +F + +M
Sbjct: 286 HFSQH-IFGASVGLQGKRWQWWWFALTYSIWKHRNSIIFSNANFDAHKLM 140
>CO979823
Length = 853
Score = 57.0 bits (136), Expect = 8e-08
Identities = 44/171 (25%), Positives = 66/171 (37%), Gaps = 13/171 (7%)
Frame = -2
Query: 1556 EDRILWREASDDIYSASSAYQFL---NEDGLPETGFWKLIWKASAPEKVRFFLWLVGRDA 1612
ED+ LW+ YS S Y L + + + F + IWK P K F W + RD
Sbjct: 705 EDKWLWKPEPGGHYSTKSGYHVLWGELTEEIQDADFAE-IWKLKIPTKAAVFAWRLVRDR 529
Query: 1613 LPTNAK-RHRHHLATTASCIRCGAVVEDAEHVLRRCPGS--VWVRGQLGRLLPWISDSAT 1669
LPT + R R + C C + E A H+ C + +W L
Sbjct: 528 LPTKSNLRRRQVMVQDMVCPLCNNIEEGAAHLFFNCTKTLPLWWESMSWVNLKTAMPQTP 349
Query: 1670 FRHWL-------FGHLQNRDNTLFCAMAWNLWKWRNSFVFEQVPWQPGDVM 1713
+H+L G R + A+ W +W+ RN VF+ + V+
Sbjct: 348 RQHFLQYGTDIADGLKSKRWKCWWIALTWTIWQHRNKVVFQNATFHGNKVL 196
>CA851030
Length = 358
Score = 56.2 bits (134), Expect = 1e-07
Identities = 25/73 (34%), Positives = 43/73 (58%)
Frame = +1
Query: 880 LQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCT 939
+++ +L QEE W Q+SR W + D+NT+FFH +T+ R+ N I + ++GN C
Sbjct: 139 VEESLDSLLKQEEEYWQQRSRAIWFKERDKNTEFFHRKTLQRKASNSITKIQDDEGNVCF 318
Query: 940 DPSTLQEEAKRFY 952
P +++ +FY
Sbjct: 319 KPDQIEDILIKFY 357
>TC211809
Length = 895
Score = 56.2 bits (134), Expect = 1e-07
Identities = 45/162 (27%), Positives = 61/162 (36%), Gaps = 17/162 (10%)
Frame = +2
Query: 1569 YSASSAYQFLNE--DGLPETGFWKLIWKASAPEKVRFFLWLVGRDALPTNAK-RHRHHLA 1625
YS SAY+ L E P + +W P K F W + +D LPT R R
Sbjct: 206 YSTKSAYRLLMEATGEFPVDRTYVDLWNLKIPSKAIVFAWRLIKDRLPTWTNLRVRQVEL 385
Query: 1626 TTASCIRCGAVVEDAEHVLRRCPGS--VWVRGQLGRLLPWISDSATFRHWLFGHLQNRDN 1683
+ C C + EDA H+ C + +W Q W++ F H H DN
Sbjct: 386 NDSRCPLCNSSEEDAAHLFFHCTKTEXLWWETQ-----SWVNSLGAFPHNPKDHFLQHDN 550
Query: 1684 T------------LFCAMAWNLWKWRNSFVFEQVPWQPGDVM 1713
L+ A+ W +WK RN VF + VM
Sbjct: 551 RSAGGVXARRWKCLWVALTWVIWKHRNGVVFHNHSFDGSKVM 676
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.336 0.145 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 96,844,662
Number of Sequences: 63676
Number of extensions: 1525760
Number of successful extensions: 11900
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 11466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11849
length of query: 1904
length of database: 12,639,632
effective HSP length: 111
effective length of query: 1793
effective length of database: 5,571,596
effective search space: 9989871628
effective search space used: 9989871628
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0228a.4


