

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
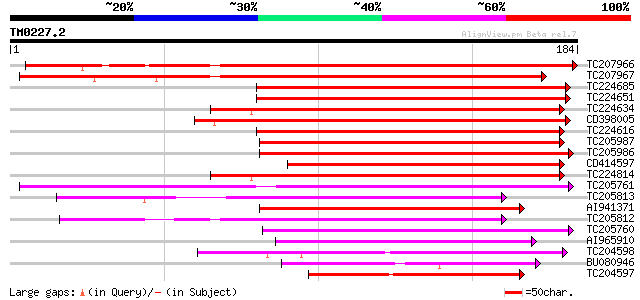
Query= TM0227.2
(184 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207966 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-... 229 6e-61
TC207967 similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,... 223 4e-59
TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 121 2e-28
TC224651 weakly similar to UP|THIM_BRANA (Q9XGS0) Thioredoxin M-... 119 6e-28
TC224634 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 118 1e-27
CD398005 118 2e-27
TC224616 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 117 3e-27
TC205987 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m... 112 1e-25
TC205986 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m... 111 2e-25
CD414597 108 1e-24
TC224814 weakly similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-... 89 1e-18
TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 88 2e-18
TC205813 weakly similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, par... 82 1e-16
AI941371 similar to GP|1388088|gb|A thioredoxin m {Pisum sativum... 82 1e-16
TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%) 79 9e-16
TC205760 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 74 3e-14
AI965910 74 5e-14
TC204598 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%) 65 1e-11
BU080946 weakly similar to GP|25990392|gb thioredoxin h {Brassic... 61 3e-10
TC204597 homologue to UP|Q9M5J3 (Q9M5J3) Beta galactosidase, par... 60 8e-10
>TC207966 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,
chloroplast precursor (TRX-M3), partial (61%)
Length = 756
Score = 229 bits (584), Expect = 6e-61
Identities = 121/180 (67%), Positives = 144/180 (79%), Gaps = 1/180 (0%)
Frame = +1
Query: 6 TSISLTSSSSSSSSSSS-SSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHL 64
TS+SL +S S +SS + + + DP SSS SH P S PF LR+ H+P Q P L
Sbjct: 1 TSLSLMASFSITSSPPTLHKATITDP--SSSSSHAFPHRS-PFVLRMPHQPHRPQTPLVL 171
Query: 65 PRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEY 124
P KI+SLR ETRA P+TKDLW+NSILKS+IPVLV FYA+WCGPCRMVHR+ID+IA EY
Sbjct: 172 P---KIQSLRNETRAIPVTKDLWDNSILKSEIPVLVIFYANWCGPCRMVHRIIDEIATEY 342
Query: 125 AGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP 184
AG+L CF+VNTDTDMQIA+DYEIKAVPVV++FK+G+KCD+VIGTMPKEFYV AIE VLKP
Sbjct: 343 AGKLKCFIVNTDTDMQIAEDYEIKAVPVVLMFKNGKKCDSVIGTMPKEFYVAAIEGVLKP 522
>TC207967 similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3, chloroplast
precursor (TRX-M3), partial (56%)
Length = 528
Score = 223 bits (568), Expect = 4e-59
Identities = 114/178 (64%), Positives = 142/178 (79%), Gaps = 7/178 (3%)
Frame = +2
Query: 4 ISTSISLTSSSSSSSSSSSSSSH---LHDPFLSSSRSHLNPSLSFP----FHLRITHKPL 56
++ S+SL+S +S S +SSS + H + DP SSS S + S +FP F LR+ H+P
Sbjct: 2 LNLSLSLSSMASFSLTSSSPTLHKATIIDPSSSSSSSSSSSSHAFPHRPPFVLRMPHQPR 181
Query: 57 HLQNPPHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRV 116
LQ P LP KI+SLR +TRATP+TKDLW+NSILKS+ PVLV FYA+WCGPCRMVHR+
Sbjct: 182 RLQTPLVLP---KIQSLRHKTRATPVTKDLWDNSILKSETPVLVIFYANWCGPCRMVHRI 352
Query: 117 IDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFY 174
ID+IA EYAG+L CF+VNTDTDMQIA+DYEIKAVPVV++FK+G+KCD+VIGTMPKEFY
Sbjct: 353 IDEIATEYAGKLKCFIVNTDTDMQIAEDYEIKAVPVVLMFKNGEKCDSVIGTMPKEFY 526
>TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1169
Score = 121 bits (304), Expect = 2e-28
Identities = 52/102 (50%), Positives = 75/102 (72%)
Frame = +3
Query: 81 PITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQ 140
PIT W++ +L+S+ PVLVEF+A WCGPCRM+H +ID++AKEY GRL C+ +NTD
Sbjct: 489 PITDANWQSLVLESESPVLVEFWAPWCGPCRMIHPIIDELAKEYVGRLKCYKLNTDESPS 668
Query: 141 IADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
A Y I+++P V++FK+G+K D VIG +PK ++IE+ L
Sbjct: 669 TATRYGIRSIPTVIIFKNGEKKDTVIGAVPKTTLTSSIEKFL 794
>TC224651 weakly similar to UP|THIM_BRANA (Q9XGS0) Thioredoxin M-type,
chloroplast precursor (TRX-M), partial (67%)
Length = 848
Score = 119 bits (299), Expect = 6e-28
Identities = 49/102 (48%), Positives = 75/102 (73%)
Frame = +2
Query: 81 PITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQ 140
P+T W++ +++S+ PVLVEF+A WCGPCRM+H +ID++AKEY G+L C+ +NTD
Sbjct: 305 PVTDANWQSLVIESESPVLVEFWAPWCGPCRMIHPIIDELAKEYTGKLKCYKLNTDESPS 484
Query: 141 IADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
A Y I+++P V++FK+G+K D VIG +PK ++IE+ L
Sbjct: 485 TATKYGIRSIPTVIIFKNGEKKDTVIGAVPKTTLTSSIEKFL 610
>TC224634 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (60%)
Length = 743
Score = 118 bits (296), Expect = 1e-27
Identities = 53/117 (45%), Positives = 79/117 (67%), Gaps = 2/117 (1%)
Frame = +2
Query: 66 RSHKIRSLRQET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKE 123
R+ ++ Q+T PIT W++ +L+S+ VLVEF+A WCGPCRM+H +ID++AK+
Sbjct: 212 RAARVACEAQDTAVEVAPITDANWQSLVLESESAVLVEFWAPWCGPCRMIHPIIDELAKQ 391
Query: 124 YAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
YAG+L C+ +NTD A Y I+++P VM+FK G+K D VIG +PK +IE+
Sbjct: 392 YAGKLKCYKLNTDESPSTATRYGIRSIPTVMIFKSGEKKDTVIGAVPKSTLTTSIEK 562
>CD398005
Length = 693
Score = 118 bits (295), Expect = 2e-27
Identities = 54/123 (43%), Positives = 80/123 (64%), Gaps = 1/123 (0%)
Frame = -3
Query: 61 PPHLP-RSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDD 119
PP P R K + P+T W + +++S+ PVLVEF+A WCGPCRM+H +ID+
Sbjct: 592 PPIGPKRGVKCEAGDTAVEVAPVTDANWLSLVIESESPVLVEFWAPWCGPCRMIHPIIDE 413
Query: 120 IAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIE 179
+AKEY G+L C+ +NTD A Y I+++P V++FK+G+K D VIG +PK ++IE
Sbjct: 412 LAKEYTGKLKCYKLNTDESPSTATKYGIRSIPTVIIFKNGEKKDTVIGAVPKTTLTSSIE 233
Query: 180 RVL 182
+ L
Sbjct: 232 KFL 224
>TC224616 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (58%)
Length = 869
Score = 117 bits (293), Expect = 3e-27
Identities = 50/100 (50%), Positives = 73/100 (73%)
Frame = +3
Query: 81 PITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQ 140
PIT W++ +L+S+ VLVEF+A WCGPCRM+H +ID++AK+YAG+L C+ +NTD
Sbjct: 333 PITDANWQSLVLESESAVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDESPS 512
Query: 141 IADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
A Y I+++P VM+FK+G+K D VIG +PK +IE+
Sbjct: 513 TATRYGIRSIPTVMIFKNGEKKDTVIGAVPKSTLTASIEK 632
>TC205987 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (64%)
Length = 891
Score = 112 bits (279), Expect = 1e-25
Identities = 46/99 (46%), Positives = 69/99 (69%)
Frame = +1
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T W N ++ S+ PVLVEF+A WCGPCRM+ ID++AKEYAG++ CF +NTD I
Sbjct: 307 VTDSSWNNLVIASETPVLVEFWAPWCGPCRMIAPAIDELAKEYAGKIACFKLNTDDSPNI 486
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
A Y I+++P V+ FK+G+K +++IG +PK +E+
Sbjct: 487 ATQYGIRSIPTVLFFKNGEKKESIIGAVPKSTLSATVEK 603
>TC205986 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (64%)
Length = 912
Score = 111 bits (278), Expect = 2e-25
Identities = 45/102 (44%), Positives = 72/102 (70%)
Frame = +1
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T W N ++ S+ PVLVEF+A WCGPCRM+ VID++AK+YAG++ C+ +NTD I
Sbjct: 319 VTDSSWNNLVIASETPVLVEFWAPWCGPCRMIAPVIDELAKDYAGKIACYKLNTDDSPNI 498
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
A Y I+++P V+ FK+G+K +++IG +PK +E+ ++
Sbjct: 499 ATQYGIRSIPTVLFFKNGEKKESIIGAVPKSTLSATVEKYVE 624
>CD414597
Length = 663
Score = 108 bits (270), Expect = 1e-24
Identities = 46/90 (51%), Positives = 67/90 (74%)
Frame = -2
Query: 91 ILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAV 150
+L+S+ VLVEF+A WCGPCRM+H +ID++AK+YAG+L C+ +NTD A Y I+++
Sbjct: 515 LLESESAVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDESPSTATRYGIRSI 336
Query: 151 PVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
P VM+FK+G+K D VIG +PK +IE+
Sbjct: 335 PTVMIFKNGEKKDTVIGAVPKSTLTASIEK 246
>TC224814 weakly similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,
chloroplast precursor (TRX-M4), partial (47%)
Length = 560
Score = 89.0 bits (219), Expect = 1e-18
Identities = 42/117 (35%), Positives = 72/117 (60%), Gaps = 2/117 (1%)
Frame = +3
Query: 66 RSHKIRSLRQET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKE 123
R+ ++ Q+T PIT W++ +L+S+ VLVEF+A WCGPCRM+H +ID++AKE
Sbjct: 210 RAARVACEAQDTAVEVAPITDANWQSLVLESESAVLVEFWAPWCGPCRMIHPIIDELAKE 389
Query: 124 YAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
GRL C+ + TD Y +++ + K+G+K D ++G++ + + +A E+
Sbjct: 390 *VGRL*CYSLTTDESPSTTTSYRNRSLLSDINIKNGEKQDRIVGSVQQITFSSAYEK 560
>TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 1023
Score = 88.2 bits (217), Expect = 2e-18
Identities = 50/180 (27%), Positives = 94/180 (51%)
Frame = +1
Query: 4 ISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPH 63
+ ++S+++SS+ +S +S S+ L +S+S S SL F R+ P
Sbjct: 178 VPMAVSVSASSAIASLNSERSTRLAPSLVSASSSAKQSSLQFTSPTRLLRISTPRAPAPS 357
Query: 64 LPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKE 123
PR + +++T + +E+ + S+ PVLV+FYA+WCGPC+ + ++++++
Sbjct: 358 RPRFLPLVQAKKQTYNS------FEDLLANSEKPVLVDFYATWCGPCQFMVPILNEVSTR 519
Query: 124 YAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
++ ++T+ IAD Y I+A+P ++FKDG D G + + + IE LK
Sbjct: 520 LQDKIQVVKIDTEKYPTIADKYRIEALPTFIMFKDGDPYDRFEGALTADQLIERIEAGLK 699
>TC205813 weakly similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (63%)
Length = 576
Score = 82.4 bits (202), Expect = 1e-16
Identities = 47/149 (31%), Positives = 82/149 (54%), Gaps = 3/149 (2%)
Frame = +1
Query: 16 SSSSSSSSSSHLHDPFLSSSRSHLNPS---LSFPFHLRITHKPLHLQNPPHLPRSHKIRS 72
S+S+S+SSS L + S+R L+ LSF H K +
Sbjct: 97 SASASTSSSVPLTHSYSCSARQRLSSHRTRLSFRNHSSAVPK----------------LT 228
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+ T I + +++++LK++ PVLVEF A+WCGPCR++ ++ +AKEY RL
Sbjct: 229 VTCGAAVTEINETQFKDTVLKANRPVLVEFVATWCGPCRLISSAMESLAKEYEDRLTVVK 408
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQK 161
++ D + Q+ ++Y++ +P ++LFK+GQ+
Sbjct: 409 IDHDANPQLIEEYKVYGLPTLILFKNGQE 495
>AI941371 similar to GP|1388088|gb|A thioredoxin m {Pisum sativum}, partial
(56%)
Length = 292
Score = 82.4 bits (202), Expect = 1e-16
Identities = 34/86 (39%), Positives = 54/86 (62%)
Frame = +2
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T W N ++ S+ PVL+EF+A CGPC M+ ID++A EYA + C +NTD + I
Sbjct: 32 VTDSSWNNLVIASETPVLLEFWAP*CGPCTMIAPAIDELANEYARTIACCKLNTDDSLNI 211
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIG 167
A Y I+ +P V+ F + + +++IG
Sbjct: 212 ASQYGIRRIPTVLFFTNAEXTESIIG 289
>TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%)
Length = 904
Score = 79.3 bits (194), Expect = 9e-16
Identities = 42/145 (28%), Positives = 84/145 (56%)
Frame = +1
Query: 17 SSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQE 76
+S+S+SSS L + +++R L+ + +P+ +P K+ +
Sbjct: 226 ASASTSSSVSLTHSWSATARQRLSSHRT---------RPIFRNYSSAVP---KLTVVTCG 369
Query: 77 TRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTD 136
T I + +++++LK++ PVLVEF A+WCGPCR++ ++ +AKEY RL ++ D
Sbjct: 370 AAVTEINETQFKDTVLKANRPVLVEFVATWCGPCRLISPSMESLAKEYEDRLTVVKIDHD 549
Query: 137 TDMQIADDYEIKAVPVVMLFKDGQK 161
+ ++ ++Y++ +P ++LFK+GQ+
Sbjct: 550 ANPRLIEEYKVYGLPTLILFKNGQE 624
>TC205760 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 666
Score = 74.3 bits (181), Expect = 3e-14
Identities = 32/101 (31%), Positives = 61/101 (59%)
Frame = +2
Query: 83 TKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIA 142
T + +++ + S+ PVLV+FYA+WCGPC+ + ++++++ ++ ++T+ IA
Sbjct: 83 TYNSFDDLLANSEKPVLVDFYATWCGPCQFMVPILNEVSTRLKDKIQVVKIDTEKYPSIA 262
Query: 143 DDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
D Y I+A+P ++FKDG+ D G + + + IE LK
Sbjct: 263 DKYRIEALPTFIMFKDGEPYDRFEGALTADQLIERIEAGLK 385
>AI965910
Length = 286
Score = 73.6 bits (179), Expect = 5e-14
Identities = 32/85 (37%), Positives = 48/85 (55%)
Frame = +2
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W++ + S+ PVL E +A WCGPCRM+H +ID++ EY L + + TD A Y
Sbjct: 26 WQSPAIASEPPVLRETWAPWCGPCRMIHPIIDELVNEYTASL*RYKLITDESPPTATKYG 205
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPK 171
I+++P +F +G IG PK
Sbjct: 206 IRSIPTANIFTNGDNTAIAIGAEPK 280
>TC204598 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%)
Length = 610
Score = 65.5 bits (158), Expect = 1e-11
Identities = 36/124 (29%), Positives = 66/124 (53%), Gaps = 4/124 (3%)
Frame = +2
Query: 62 PHLPRSHKIRSLRQETRATPI-TKDLWENSILK---SDIPVLVEFYASWCGPCRMVHRVI 117
P +S +R++ +E + + T D W + S ++V+F ASWCGPCR + V+
Sbjct: 23 PGHSQSGSLRNMAEEGQVVGVHTVDAWNQQLQNGKDSQKLIVVDFTASWCGPCRFIAPVL 202
Query: 118 DDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNA 177
+IA+ + ++ V+ D +A++Y I+A+P + KDG+ D V+G +E +
Sbjct: 203 AEIAR-HTPQVIFLKVDVDEVRPVAEEYSIEAMPTFLFLKDGKIVDKVVGAKKEELQLTI 379
Query: 178 IERV 181
+ V
Sbjct: 380 AKHV 391
>BU080946 weakly similar to GP|25990392|gb thioredoxin h {Brassica rapa},
partial (76%)
Length = 409
Score = 61.2 bits (147), Expect = 3e-10
Identities = 29/86 (33%), Positives = 52/86 (59%), Gaps = 2/86 (2%)
Frame = +3
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD--MQIADDYE 146
N + ++ V+++F ASWCGPCR + V +++AK+++ N V D D +A D++
Sbjct: 120 NEVKETSKLVVIDFTASWCGPCRFIAPVFNEMAKKFS---NAEFVKIDVDELPDVAKDFK 290
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKE 172
++A+P +L K G++ D V+G E
Sbjct: 291 VEAMPTFVLCKKGKEVDRVVGARKDE 368
>TC204597 homologue to UP|Q9M5J3 (Q9M5J3) Beta galactosidase, partial (14%)
Length = 1372
Score = 59.7 bits (143), Expect = 8e-10
Identities = 28/70 (40%), Positives = 44/70 (62%)
Frame = +1
Query: 98 VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFK 157
++V+F ASWCGPCR + V+ +IAK+ L V+ D +A++Y I+A+P + K
Sbjct: 172 IVVDFTASWCGPCRFMAPVLAEIAKK-TPELIFLKVDVDEVRPVAEEYSIEAMPTFLFLK 348
Query: 158 DGQKCDAVIG 167
DG+ D V+G
Sbjct: 349 DGEIVDKVVG 378
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,623,227
Number of Sequences: 63676
Number of extensions: 237050
Number of successful extensions: 10115
Number of sequences better than 10.0: 518
Number of HSP's better than 10.0 without gapping: 4007
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6750
length of query: 184
length of database: 12,639,632
effective HSP length: 91
effective length of query: 93
effective length of database: 6,845,116
effective search space: 636595788
effective search space used: 636595788
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0227.2


