

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
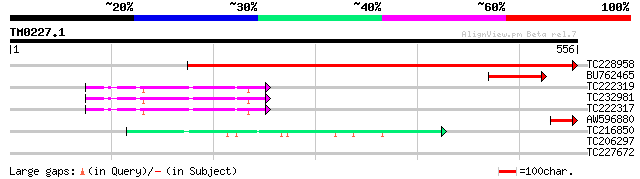
Query= TM0227.1
(556 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228958 similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-CoA ca... 723 0.0
BU762465 104 9e-23
TC222319 UP|Q39833 (Q39833) Alfa-carboxyltransferase precursor (... 51 1e-06
TC232981 UP|Q9LLR0 (Q9LLR0) Carboxyl transferase alpha subunit ... 51 1e-06
TC222317 UP|Q9LLQ8 (Q9LLQ8) Carboxyl transferase alpha subunit ... 51 1e-06
AW596880 46 5e-05
TC216850 homologue to UP|Q42793 (Q42793) Acetyl CoA carboxylase ... 44 2e-04
TC206297 UP|Q8GZD3 (Q8GZD3) Expansin, complete 30 2.8
TC227672 weakly similar to UP|Q9LKG1 (Q9LKG1) Histone deacetylas... 28 8.2
>TC228958 similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-CoA carboxylase
beta chain, mitochondrial precursor
(3-Methylcrotonyl-CoA carboxylase 2) (MCCase beta
subunit) (3-methylcrotonyl-CoA:carbon dioxide ligase
beta subunit) , partial (63%)
Length = 1226
Score = 723 bits (1867), Expect = 0.0
Identities = 358/382 (93%), Positives = 375/382 (97%)
Frame = +2
Query: 175 RENFGRIFYNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLV 234
RENFGRIFYNQA+MSA+GIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLV
Sbjct: 2 RENFGRIFYNQAVMSAQGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLV 181
Query: 235 KAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLHMAGKDVLANGLQ 294
KAATGEEVSAEDLGGATVHCKTSGVSDYFAQDE HALALGRNIIKNLHMAGKDVLANGLQ
Sbjct: 182 KAATGEEVSAEDLGGATVHCKTSGVSDYFAQDELHALALGRNIIKNLHMAGKDVLANGLQ 361
Query: 295 NLSYEYKEPLYDVNELRSIAPTDLKKQFDVRSVIARIVDGSEFDEFKKLYGTTLVTGFAR 354
N++YEYKEPLYD+NELRSIAPTDLK+QFD+RSVI RIVDGSEFDEFKKLYGTTLVTGFAR
Sbjct: 362 NINYEYKEPLYDINELRSIAPTDLKQQFDIRSVIDRIVDGSEFDEFKKLYGTTLVTGFAR 541
Query: 355 IYGQPVGIIGNNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMVGSRSEANGIAKS 414
I+GQPVGIIGNNGILFNESALKGAHFIE+CTQRNIPLVFLQNITGFMVGSRSEANGIAKS
Sbjct: 542 IFGQPVGIIGNNGILFNESALKGAHFIELCTQRNIPLVFLQNITGFMVGSRSEANGIAKS 721
Query: 415 GAKMVMAVSCAKVPKITIIVGGSFGAGNYAMCGRAYSPNFMFLWPNARISVMGGAQAAGV 474
GAKMVMAVSCAKVPK+TI+VGGSFGAGNYAMCGRAYSPNF+FLWPNARISVMGGAQAAGV
Sbjct: 722 GAKMVMAVSCAKVPKVTIMVGGSFGAGNYAMCGRAYSPNFLFLWPNARISVMGGAQAAGV 901
Query: 475 LSQIEKTNKKKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWDDGIIDPADTRKVI 534
L+QIEK NKK+QGIQWSKEEE+KFK KVVEAYE E SPYYSTARLWDDGIIDPADTRKVI
Sbjct: 902 LAQIEKGNKKRQGIQWSKEEEDKFKGKVVEAYEREASPYYSTARLWDDGIIDPADTRKVI 1081
Query: 535 GLSISASLNRAIEDTKFGVFRM 556
GL ISASLNRAIE+TK+GVFRM
Sbjct: 1082GLCISASLNRAIENTKYGVFRM 1147
>BU762465
Length = 429
Score = 104 bits (260), Expect = 9e-23
Identities = 50/57 (87%), Positives = 53/57 (92%)
Frame = +3
Query: 470 QAAGVLSQIEKTNKKKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWDDGIID 526
QAAGVL+QIEK NKK+QGIQWSKEEE+KFK KVVEAYE E SPYYSTARLWDDGIID
Sbjct: 258 QAAGVLAQIEKGNKKRQGIQWSKEEEDKFKGKVVEAYEREASPYYSTARLWDDGIID 428
>TC222319 UP|Q39833 (Q39833) Alfa-carboxyltransferase precursor (Carboxyl
transferase alpha subunit) , complete
Length = 2588
Score = 51.2 bits (121), Expect = 1e-06
Identities = 56/199 (28%), Positives = 88/199 (44%), Gaps = 18/199 (9%)
Frame = +2
Query: 75 IDRLLDPGSSFLEL-SQLAGHELYEEPLPSGGVVTGIGPVHGRLCMFVANDPTVKG---- 129
+D + + F+EL AG Y++P +VTG+G + GR MF+ + KG
Sbjct: 827 LDHVFNITEKFVELHGDRAG---YDDP----AIVTGLGTIDGRSYMFIGHQ---KGRNTK 976
Query: 130 -------GTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIF 182
G P +K LR E A P V +D+ GA+ +++ E I
Sbjct: 977 ENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYADLKSEELGQGE---AIA 1147
Query: 183 YNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPP-----LVKAA 237
+N M +P I++V+G +GGA A++ +M++ N ++A P L K A
Sbjct: 1148 HNLRSMFGLKVPVISIVIGEGGSGGALAIGCANKLLMLE-NAVFYVASPEACAAILWKTA 1324
Query: 238 TGEEVSAEDLG-GATVHCK 255
+AE L AT CK
Sbjct: 1325 KASPKAAEKLKITATELCK 1381
Score = 33.5 bits (75), Expect = 0.26
Identities = 47/202 (23%), Positives = 76/202 (37%), Gaps = 15/202 (7%)
Frame = +2
Query: 298 YEYKEPLYDVNELRSI-APTDLKKQFDVRSVIARIV-DGSEFDEFKKLYGTTLVTGFARI 355
Y + P+ VN R PT L F++ + D + +D+ +VTG I
Sbjct: 764 YTHLTPIQRVNIARHPNRPTFLDHVFNITEKFVELHGDRAGYDD------PAIVTGLGTI 925
Query: 356 YGQPVGIIG-------------NNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV 402
G+ IG N G+ K +E P+V + G
Sbjct: 926 DGRSYMFIGHQKGRNTKENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYA 1105
Query: 403 GSRSEANGIAKSGAKMVMAVSCAKVPKITIIVGGSFGAGNYAMCGRAYSPNFMFLWPNAR 462
+SE G ++ A + ++ KVP I+I++G G+G G A N + + NA
Sbjct: 1106DLKSEELGQGEAIAHNLRSMFGLKVPVISIVIGEG-GSGGALAIGCA---NKLLMLENAV 1273
Query: 463 ISVMGGAQAAGVLSQIEKTNKK 484
V A +L + K + K
Sbjct: 1274FYVASPEACAAILWKTAKASPK 1339
>TC232981 UP|Q9LLR0 (Q9LLR0) Carboxyl transferase alpha subunit , complete
Length = 2799
Score = 51.2 bits (121), Expect = 1e-06
Identities = 56/199 (28%), Positives = 88/199 (44%), Gaps = 18/199 (9%)
Frame = +3
Query: 75 IDRLLDPGSSFLEL-SQLAGHELYEEPLPSGGVVTGIGPVHGRLCMFVANDPTVKG---- 129
+D + + F+EL AG Y++P +VTG+G + GR MF+ + KG
Sbjct: 819 LDHVFNITEKFVELHGDRAG---YDDP----AIVTGLGTIDGRSYMFIGHQ---KGRNTK 968
Query: 130 -------GTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIF 182
G P +K LR E A P V +D+ GA+ +++ E I
Sbjct: 969 ENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYADLKSEELGQGE---AIA 1139
Query: 183 YNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPP-----LVKAA 237
+N M +P I++V+G +GGA A++ +M++ N ++A P L K A
Sbjct: 1140 HNLRSMFGLKVPVISIVIGEGGSGGALAIGCANKLLMLE-NAVFYVASPEACAAILWKTA 1316
Query: 238 TGEEVSAEDLG-GATVHCK 255
+AE L AT CK
Sbjct: 1317 KASPKAAEKLKITATELCK 1373
Score = 33.5 bits (75), Expect = 0.26
Identities = 47/202 (23%), Positives = 76/202 (37%), Gaps = 15/202 (7%)
Frame = +3
Query: 298 YEYKEPLYDVNELRSI-APTDLKKQFDVRSVIARIV-DGSEFDEFKKLYGTTLVTGFARI 355
Y + P+ VN R PT L F++ + D + +D+ +VTG I
Sbjct: 756 YTHLTPIQRVNIARHPNRPTFLDHVFNITEKFVELHGDRAGYDD------PAIVTGLGTI 917
Query: 356 YGQPVGIIG-------------NNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV 402
G+ IG N G+ K +E P+V + G
Sbjct: 918 DGRSYMFIGHQKGRNTKENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYA 1097
Query: 403 GSRSEANGIAKSGAKMVMAVSCAKVPKITIIVGGSFGAGNYAMCGRAYSPNFMFLWPNAR 462
+SE G ++ A + ++ KVP I+I++G G+G G A N + + NA
Sbjct: 1098DLKSEELGQGEAIAHNLRSMFGLKVPVISIVIGEG-GSGGALAIGCA---NKLLMLENAV 1265
Query: 463 ISVMGGAQAAGVLSQIEKTNKK 484
V A +L + K + K
Sbjct: 1266FYVASPEACAAILWKTAKASPK 1331
>TC222317 UP|Q9LLQ8 (Q9LLQ8) Carboxyl transferase alpha subunit , complete
Length = 2521
Score = 51.2 bits (121), Expect = 1e-06
Identities = 56/199 (28%), Positives = 88/199 (44%), Gaps = 18/199 (9%)
Frame = +3
Query: 75 IDRLLDPGSSFLEL-SQLAGHELYEEPLPSGGVVTGIGPVHGRLCMFVANDPTVKG---- 129
+D + + F+EL AG Y++P +VTG+G + GR MF+ + KG
Sbjct: 846 LDHVFNITEKFVELHGDRAG---YDDP----AIVTGLGTIDGRSYMFIGHQ---KGRNTK 995
Query: 130 -------GTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIF 182
G P +K LR E A P V +D+ GA+ +++ E I
Sbjct: 996 ENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYADLKSEELGQGE---AIA 1166
Query: 183 YNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPP-----LVKAA 237
+N M +P I++V+G +GGA A++ +M++ N ++A P L K A
Sbjct: 1167 HNLRSMFGLKVPVISIVIGEGGSGGALAIGCANKLLMLE-NAVFYVASPEACAAILWKTA 1343
Query: 238 TGEEVSAEDLG-GATVHCK 255
+AE L AT CK
Sbjct: 1344 KASPKAAEKLKITATELCK 1400
Score = 33.5 bits (75), Expect = 0.26
Identities = 47/202 (23%), Positives = 76/202 (37%), Gaps = 15/202 (7%)
Frame = +3
Query: 298 YEYKEPLYDVNELRSI-APTDLKKQFDVRSVIARIV-DGSEFDEFKKLYGTTLVTGFARI 355
Y + P+ VN R PT L F++ + D + +D+ +VTG I
Sbjct: 783 YTHLTPIQRVNIARHPNRPTFLDHVFNITEKFVELHGDRAGYDD------PAIVTGLGTI 944
Query: 356 YGQPVGIIG-------------NNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV 402
G+ IG N G+ K +E P+V + G
Sbjct: 945 DGRSYMFIGHQKGRNTKENIQRNFGMPTPHGYRKALRLMEYADHHGFPIVTFIDTPGAYA 1124
Query: 403 GSRSEANGIAKSGAKMVMAVSCAKVPKITIIVGGSFGAGNYAMCGRAYSPNFMFLWPNAR 462
+SE G ++ A + ++ KVP I+I++G G+G G A N + + NA
Sbjct: 1125DLKSEELGQGEAIAHNLRSMFGLKVPVISIVIGEG-GSGGALAIGCA---NKLLMLENAV 1292
Query: 463 ISVMGGAQAAGVLSQIEKTNKK 484
V A +L + K + K
Sbjct: 1293FYVASPEACAAILWKTAKASPK 1358
>AW596880
Length = 123
Score = 45.8 bits (107), Expect = 5e-05
Identities = 22/26 (84%), Positives = 25/26 (95%)
Frame = +2
Query: 531 RKVIGLSISASLNRAIEDTKFGVFRM 556
RKVIGLSISASLNR I++TK+GVFRM
Sbjct: 2 RKVIGLSISASLNRDIQNTKYGVFRM 79
>TC216850 homologue to UP|Q42793 (Q42793) Acetyl CoA carboxylase , complete
Length = 7100
Score = 43.5 bits (101), Expect = 2e-04
Identities = 87/405 (21%), Positives = 149/405 (36%), Gaps = 91/405 (22%)
Frame = +1
Query: 115 GRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPD 174
GR + VAND T K G++ P ++A KLP +YL + GA L +V
Sbjct: 4792 GRTILVVANDVTFKAGSFGPREDAFFRAVTDLACTKKLPLIYLAANSGARLGVAEEV--- 4962
Query: 175 RENFGRIFYNQALMSAEGIPQIALVLGSCTAGGAYIPA-----------MADESVMVK-- 221
++ R+ +++ G + L G+ + A ++++ K
Sbjct: 4963 -KSCFRVGWSEESNPENGFQYVYLTPEDNARIGSSVIAHELKLESGETRWVIDTIVGKED 5139
Query: 222 GNGTIFLAGPPLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQ---------------D 266
G G L+G + A E V +T G+ Y A+
Sbjct: 5140 GLGVENLSGSGAIAGAYSRAYK-ETFTLTYVTGRTVGIGAYLARLGMRCIQRLDQPIILT 5316
Query: 267 EFHAL--ALGRNII-KNLHMAGKDVLA-NGLQNLSY-EYKEPLYDVNELRSIAPTDL--- 318
F AL LGR + ++ + G ++A NG+ +L+ + E + + + S P+ +
Sbjct: 5317 GFSALNKLLGREVYSSHMQLGGPKIMATNGVVHLTVSDDLEGISSILKWLSYIPSHVGGA 5496
Query: 319 ------------------KKQFDVRSVIARIVDGS-----------EFDEFKKLYGTTLV 349
+ D R+ I+ +DG+ F E + + T+V
Sbjct: 5497 LPIVKPLDPPERPVEYFPENSCDPRAAISGTLDGNGRWLGGIFDKDSFVETLEGWARTVV 5676
Query: 350 TGFARIYGQPVGIIG--------------------------NNGILFNESALKGAHFIEI 383
TG A++ G PVG++ + F +SA K A I
Sbjct: 5677 TGRAKLGGIPVGVVAVETQTVMQIIPADPGQLDSHERVVPQAGQVWFPDSATKTAQAILD 5856
Query: 384 CTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCAKVP 428
+ +PL L N GF G R GI ++G+ +V + K P
Sbjct: 5857 FNREELPLFILANWRGFSGGQRDLFEGILQAGSTIVENLRTYKQP 5991
>TC206297 UP|Q8GZD3 (Q8GZD3) Expansin, complete
Length = 1262
Score = 30.0 bits (66), Expect = 2.8
Identities = 15/50 (30%), Positives = 27/50 (54%)
Frame = +1
Query: 426 KVPKITIIVGGSFGAGNYAMCGRAYSPNFMFLWPNARISVMGGAQAAGVL 475
K+ KI +++G G + + A+SP+ W NA + GG+ A+G +
Sbjct: 217 KMGKIMLVLGSLIGLCCFTITTYAFSPSG---WTNAHATFYGGSDASGTM 357
>TC227672 weakly similar to UP|Q9LKG1 (Q9LKG1) Histone deacetylase, partial
(15%)
Length = 1420
Score = 28.5 bits (62), Expect = 8.2
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 17/111 (15%)
Frame = +3
Query: 344 YGTTLVTGFARIYGQPVGIIGNNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMVG 403
YG +L G + + P+ N ++F +K AH I ++ + L I G + G
Sbjct: 636 YGRSLCQGSSSLL-DPLR*SNNIHLVFLGKMIKQAHAFTINLVCHLSIAVLSGI-GPLQG 809
Query: 404 SRS-----------------EANGIAKSGAKMVMAVSCAKVPKITIIVGGS 437
R E ++KS + V SC K+PKIT++ GGS
Sbjct: 810 DRCISVQWCVRPLRLYPRVVERQIVSKSSRRSV*--SCFKIPKITLLSGGS 956
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,044,882
Number of Sequences: 63676
Number of extensions: 288579
Number of successful extensions: 1292
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1283
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1292
length of query: 556
length of database: 12,639,632
effective HSP length: 102
effective length of query: 454
effective length of database: 6,144,680
effective search space: 2789684720
effective search space used: 2789684720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0227.1


