

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0223.2
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
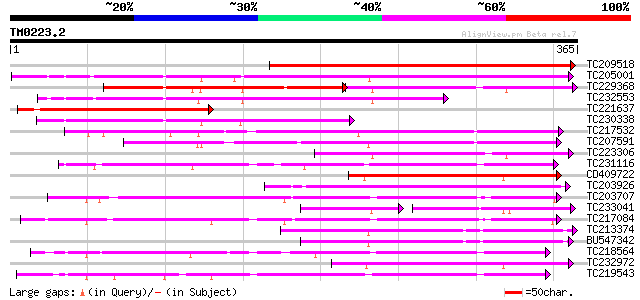
Score E
Sequences producing significant alignments: (bits) Value
TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fr... 362 e-100
TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetyles... 233 1e-61
TC229368 similar to PIR|T48618|T48618 early nodule-specific prot... 130 6e-58
TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%) 204 5e-53
TC221637 similar to UP|O82076 (O82076) Lipase homolog (Fragment)... 184 4e-47
TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%) 178 3e-45
TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 172 3e-43
TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 145 3e-35
TC223306 weakly similar to PIR|T48618|T48618 early nodule-specif... 140 9e-34
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 136 1e-32
CD409722 136 1e-32
TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H... 127 8e-30
TC203707 125 3e-29
TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment)... 80 6e-29
TC217084 120 1e-27
TC213374 weakly similar to UP|Q94F40 (Q94F40) At1g28600/F1K23_6,... 119 3e-27
BU547342 weakly similar to GP|10764858|gb| F1K23.17 {Arabidopsis... 118 4e-27
TC218564 118 4e-27
TC232972 weakly similar to UP|O82681 (O82681) Lanatoside 15'-O-a... 115 2e-26
TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase E... 115 4e-26
>TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fragment),
partial (56%)
Length = 874
Score = 362 bits (930), Expect = e-100
Identities = 170/197 (86%), Positives = 181/197 (91%)
Frame = +1
Query: 168 QNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL 227
QNDLADSFAKNL+Y QVIKKIP VITEIENAVK+LYN+GARKFWVHNTGPLGCLPK+LAL
Sbjct: 1 QNDLADSFAKNLSYAQVIKKIPAVITEIENAVKNLYNDGARKFWVHNTGPLGCLPKVLAL 180
Query: 228 AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATK 287
AQKKDLD GCLSSYNSAARLFNEAL HSSQKLR+ KDATLVYVDIYAIK+DLI NA K
Sbjct: 181 AQKKDLDSLGCLSSYNSAARLFNEALLHSSQKLRSEFKDATLVYVDIYAIKYDLITNAAK 360
Query: 288 YGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKI 347
YGFSNPL VCCG+GGPPYNFD+RVTCGQPGYQVCDEG+RYV+WDG H TEAANT IASKI
Sbjct: 361 YGFSNPLMVCCGYGGPPYNFDVRVTCGQPGYQVCDEGARYVSWDGIHQTEAANTLIASKI 540
Query: 348 LSTDYSTPRTPFEFFCH 364
LS YSTPR PF+FFCH
Sbjct: 541 LSMAYSTPRIPFDFFCH 591
>TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetylesterase
precursor, partial (62%)
Length = 1384
Score = 233 bits (593), Expect = 1e-61
Identities = 144/379 (37%), Positives = 198/379 (51%), Gaps = 17/379 (4%)
Frame = +1
Query: 2 SSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPIN-LP 60
SS FS L+ + + S + S CD A+ F FGDSNSDTGG + FP P
Sbjct: 70 SSLYIFSKFLVICMVMMISLVDS-SYSLCDFEAI-FNFGDSNSDTGGFHTS--FPAQPAP 237
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK 120
G T+F + GR SDGRL++D L Q L +L+PYL S+ GS +T+GANFA S+ +P
Sbjct: 238 YGMTYFKKPVGRASDGRLIVDFLAQGLGLPYLSPYLQSI-GSDYTHGANFASSASTVIPP 414
Query: 121 YV--------PFSLNIQVMQFLHFKARTLEL------VSAGAKNVINDEGFRAALYLIDI 166
PFSL++Q+ Q FKA+ E +S+G K + + + F ALY I
Sbjct: 415 TTSFSVSGLSPFSLSVQLRQMEQFKAKVDEFHQTGTRISSGTK-IPSPDIFGKALYTFYI 591
Query: 167 GQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILA 226
GQND A V +P ++++I A+K LY +G R F V N GP+GC P L
Sbjct: 592 GQNDFTSKIAATGGIDGVRGSLPHIVSQINAAIKELYAQGGRAFMVFNLGPVGCYPGYLV 771
Query: 227 LAQK--KDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIAN 284
D D FGC+ S+N+A +N+ L + + L DA+L+Y D ++ +L +
Sbjct: 772 ELPHATSDYDEFGCIVSHNNAVNDYNKLLRDTLTQTGESLVDASLIYADTHSALLELFHH 951
Query: 285 ATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIA 344
T YG CCG+GG YNF+ ++ CG CDE YV+WDG H TEAAN +A
Sbjct: 952 PTFYGLKYNTRTCCGYGGGVYNFNPKILCGHMLASACDEPQNYVSWDGIHFTEAANKIVA 1131
Query: 345 SKILSTDYSTPRTPFEFFC 363
IL+ P P C
Sbjct: 1132HAILNGSLFYPPFPLHKHC 1188
>TC229368 similar to PIR|T48618|T48618 early nodule-specific protein-like -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(61%)
Length = 1238
Score = 130 bits (326), Expect(2) = 6e-58
Identities = 71/170 (41%), Positives = 107/170 (62%), Gaps = 13/170 (7%)
Frame = +2
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---- 116
+G FFH+ +GR DGRL++D + + LN +L+ YL+SL G+++ +GANFA GS+
Sbjct: 2 DGEGFFHKPSGRDCDGRLIVDFIAEKLNLPYLSAYLNSL-GTNYRHGANFATGGSTIRKQ 178
Query: 117 --TLPKY--VPFSLNIQVMQFLHFKARTLELVSAGA-----KNVINDEGFRAALYLIDIG 167
T+ +Y PFSL+IQ++QF FKART +L + E F ALY DIG
Sbjct: 179 NETIFQYGISPFSLDIQIVQFNQFKARTKQLYEEAKAPHEKSKLPVPEEFSKALYTFDIG 358
Query: 168 QNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGP 217
QNDL+ F K + + Q+ + +P ++ ++ NAVK++Y +G R FW+HNT P
Sbjct: 359 QNDLSVGFRK-MNFDQIRESMPDILNQLANAVKNIYQQGGRYFWIHNTSP 505
Score = 112 bits (280), Expect(2) = 6e-58
Identities = 59/160 (36%), Positives = 86/160 (52%), Gaps = 11/160 (6%)
Frame = +3
Query: 217 PLGCLPKILALAQKKD---LDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVD 273
P GC+P L LD +GC+ N A FN+ L KLRT L +A + YVD
Sbjct: 504 PFGCMPVQLFYKHNIPEGYLDQYGCVKDQNVMATEFNKQLKDRVIKLRTELPEAAITYVD 683
Query: 274 IYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGY--------QVCDEGS 325
+YA K+ LI+N K GF +P+ +CCG+ + D + CG G C+ S
Sbjct: 684 VYAAKYALISNTKKEGFVDPMKICCGY----HVNDTHIWCGNLGTDNGKDVFGSACENPS 851
Query: 326 RYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFCHR 365
+Y++WD H+ EAAN ++A++IL+ Y+ P TP C+R
Sbjct: 852 QYISWDSVHYAEAANHWVANRILNGSYTDPPTPITQACYR 971
>TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%)
Length = 832
Score = 204 bits (519), Expect = 5e-53
Identities = 117/278 (42%), Positives = 164/278 (58%), Gaps = 14/278 (5%)
Frame = +2
Query: 19 TSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRL 78
T+S AA +C PA+ F FGDSNSDTGGL++ G P+G ++FH GR DGRL
Sbjct: 2 TNSLAA--SKQCHFPAI-FNFGDSNSDTGGLSAAFG-QAGPPHGESYFHHPAGRYCDGRL 169
Query: 79 VIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK---------YVPFSLNIQ 129
++D L + L +L+ +LDS+ GS++++GANFA GS+ P+ + PFSL++Q
Sbjct: 170 IVDFLAKKLGLPYLSAFLDSV-GSNYSHGANFATAGSTIRPQNTTLHQTGGFSPFSLDVQ 346
Query: 130 VMQFLHFKARTLELVSAGA--KNVI-NDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIK 186
QF F+ RT + G K ++ E F ALY DIGQNDLA + N++ QV
Sbjct: 347 FNQFSDFQRRTQFFHNKGGVYKTLLPKAEDFSQALYTFDIGQNDLASGYFHNMSTDQVKA 526
Query: 187 KIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKD--LDLFGCLSSYNS 244
+P V+ + +N +K +YN G R FWVHNTGP+GCLP I+ L K +D C + YN
Sbjct: 527 YVPDVLAQFKNVIKYVYNHGGRSFWVHNTGPVGCLPYIMDLHPVKPSLVDKAXCATPYNE 706
Query: 245 AARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLI 282
A+ FN L +L L A + YVD+Y++KH LI
Sbjct: 707 VAKFFNSKLKEVVVQLTKELPLAAITYVDVYSVKHSLI 820
>TC221637 similar to UP|O82076 (O82076) Lipase homolog (Fragment), partial
(30%)
Length = 443
Score = 184 bits (468), Expect = 4e-47
Identities = 95/126 (75%), Positives = 102/126 (80%)
Frame = +1
Query: 6 AFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTF 65
A S+ LL L L S + ++ CD VLFVFGDSNSDTGGL SGLGFPIN PNGR F
Sbjct: 73 AISVSLLLLLL---SCFTEIALAGCDKAPVLFVFGDSNSDTGGLASGLGFPINPPNGRNF 243
Query: 66 FHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFS 125
FHRSTGRLSDGRL+IDLLC SLN L PYLD+LSG+SFTNGANFAVVGSSTLPKYVPFS
Sbjct: 244 FHRSTGRLSDGRLLIDLLCLSLNASLLVPYLDALSGTSFTNGANFAVVGSSTLPKYVPFS 423
Query: 126 LNIQVM 131
LNIQVM
Sbjct: 424 LNIQVM 441
>TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%)
Length = 737
Score = 178 bits (452), Expect = 3e-45
Identities = 98/213 (46%), Positives = 127/213 (59%), Gaps = 8/213 (3%)
Frame = +2
Query: 18 ITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPI-NLPNGRTFFHRSTGRLSDG 76
+ + + A CD PA+ F FG SN+DTGGL + PNG T+FHR GR SDG
Sbjct: 104 LNNPAMATKQYYCDFPAI-FNFGASNADTGGLAASFFVAAPKSPNGETYFHRPAGRFSDG 280
Query: 77 RLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYV----PFSLNIQVMQ 132
RL+ID L QS +L+PYLDSL G++F+ GA+FA GS+ +P+ PFSL +Q Q
Sbjct: 281 RLIIDFLAQSFGLPYLSPYLDSL-GTNFSRGASFATAGSTIIPQQSFRSSPFSLGVQYSQ 457
Query: 133 FLHFKARTLELVSAG---AKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIP 189
F FK T + G A + +E F ALY DIGQNDL F N+T Q IP
Sbjct: 458 FQRFKPTTQFIREQGGVFATLMPKEEYFHEALYTFDIGQNDLTAGFFGNMTLQQFNATIP 637
Query: 190 TVITEIENAVKSLYNEGARKFWVHNTGPLGCLP 222
+I + +K++YN GAR FW+HNTGP+GCLP
Sbjct: 638 DIIKSFTSNIKNIYNMGARSFWIHNTGPIGCLP 736
>TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(54%)
Length = 1372
Score = 172 bits (435), Expect = 3e-43
Identities = 119/346 (34%), Positives = 174/346 (49%), Gaps = 25/346 (7%)
Frame = +1
Query: 36 LFVFGDSNSDTGGL--TSGLGFPINL--PNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRF 91
LF FGDS +DTG L S P L P G+T FHR GR SDGRL++D L +SL +
Sbjct: 121 LFSFGDSLTDTGNLYFISPRQSPDCLLPPYGQTHFHRPNGRCSDGRLILDFLAESLGLPY 300
Query: 92 LTPYLDSLSGS----SFTNGANFAVVGSSTLPK------------YVPFSLNIQVMQFLH 135
+ PYL +G+ + G NFAV G++ L + FSL +Q+ F
Sbjct: 301 VKPYLGFKNGAVKRGNIEQGVNFAVAGATALDRGFFEEKGFAVDVTANFSLGVQLDWFKE 480
Query: 136 FKARTLELVSAGAKNVINDEGFRAALYLI-DIGQNDLADSFAKNLTYVQVIKKIPTVITE 194
+L S+ K VI ++L+++ +IG ND ++ + ++ IP VI+
Sbjct: 481 LLP-SLCNSSSSCKKVIG-----SSLFIVGEIGGNDYGYPLSETTAFGDLVTYIPQVISV 642
Query: 195 IENAVKSLYNEGARKFWVHNTGPLGCLPK---ILALAQKKDLDLFGCLSSYNSAARLFNE 251
I +A++ L + GA F V + PLGC P I A K++ D GCL N+ NE
Sbjct: 643 ITSAIRELIDLGAVTFMVPGSLPLGCNPAYLTIFATIDKEEYDQAGCLKWLNTFYEYHNE 822
Query: 252 ALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFS-NPLTVCCGFGGPPYNFDLR 310
L +LR ++Y D + + + ++GF N L VCCG GG PYN++
Sbjct: 823 LLQIEINRLRVLYPLTNIIYADYFNAALEFYNSPEQFGFGGNVLKVCCG-GGGPYNYNET 999
Query: 311 VTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPR 356
CG G CD+ S+YV+WDG H TEAA ++ +L Y+ P+
Sbjct: 1000AMCGDAGVVACDDPSQYVSWDGYHLTEAAYRWMTKGLLDGPYTIPK 1137
>TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(57%)
Length = 1290
Score = 145 bits (366), Expect = 3e-35
Identities = 102/300 (34%), Positives = 146/300 (48%), Gaps = 18/300 (6%)
Frame = +1
Query: 74 SDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----------KYV- 122
SDGRL+ID + ++ + +L PYL G NFAV G++ L KY+
Sbjct: 4 SDGRLMIDFIAEAYDLPYLPPYLALTKDKDIQRGVNFAVAGATALDAKFFIEAGLAKYLW 183
Query: 123 -PFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLI-DIGQNDLA-DSFAKNL 179
SLNIQ+ F K S D F+ +L+L+ +IG ND + A N+
Sbjct: 184 TNNSLNIQLGWFKKLKP------SLCTTKQDCDSYFKRSLFLVGEIGGNDYNYAAIAGNI 345
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQ---KKDLDLF 236
T +Q +P V+ I A+ L EGAR+ V P+GC L L + K+D D
Sbjct: 346 TQLQAT--VPPVVEAITAAINELIAEGARELLVPGNFPIGCSALYLTLFRSENKEDYDES 519
Query: 237 GCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSN-PLT 295
GCL ++N A N+ L + + LR + A ++Y D Y +GF+N L
Sbjct: 520 GCLKTFNGFAEYHNKELKLALETLRKKNPHARILYADYYGAAKRFFHAPGHHGFTNGALR 699
Query: 296 VCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTP 355
CCG GG P+NF++ CG G + C + S Y NWDG H TEAA +IA ++ +S P
Sbjct: 700 ACCG-GGGPFNFNISARCGHTGSKACADPSTYANWDGIHLTEAAYRYIAKGLIYGPFSYP 876
>TC223306 weakly similar to PIR|T48618|T48618 early nodule-specific
protein-like - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (25%)
Length = 669
Score = 140 bits (353), Expect = 9e-34
Identities = 71/182 (39%), Positives = 107/182 (58%), Gaps = 15/182 (8%)
Frame = +1
Query: 197 NAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKD-------LDLFGCLSSYNSAARLF 249
N V++L GAR FW+HNTGP+GCLP + + + LD GC++ N AR F
Sbjct: 10 NQVQTLLGLGARTFWIHNTGPIGCLPVAMPVHNAMNTTPGAGYLDQNGCINYQNDMAREF 189
Query: 250 NEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDL 309
N+ L ++ KLR + DA+L+YVD+++ K++LI+NA K GF +P +CCG+ Y+
Sbjct: 190 NKKLKNTVVKLRVQFPDASLIYVDMFSAKYELISNANKEGFVDPSGICCGYHQDGYH--- 360
Query: 310 RVTCGQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEF 361
+ CG CD+ S+Y++WDG H+TEAAN +IA++IL+ +S P
Sbjct: 361 -LYCGNKAIINGKEIFADTCDDPSKYISWDGVHYTEAANHWIANRILNGSFSDPPLSIAH 537
Query: 362 FC 363
C
Sbjct: 538 SC 543
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 136 bits (343), Expect = 1e-32
Identities = 109/342 (31%), Positives = 160/342 (45%), Gaps = 20/342 (5%)
Frame = +1
Query: 32 VPAVLFVFGDSNSDTGGLTSGL-----GFPINLPNGRTFF-HRSTGRLSDGRLVIDLLCQ 85
VPA+ F FGDS D G L FP P GR F H TGR +G+L D +
Sbjct: 439 VPAI-FTFGDSIVDVGNNNHQLTIVKANFP---PYGRDFENHFPTGRFCNGKLATDFIAD 606
Query: 86 SLN-TRFLTPYLD-SLSGSSFTNGANFAVVGSS----TLPKYVPFSLNIQVMQFLHFKAR 139
L T + YL+ G + NGANFA S T Y L+ Q+ + + +
Sbjct: 607 ILGFTSYQPAYLNLKTKGKNLLNGANFASASSGYFELTSKLYSSIPLSKQLEYYKECQTK 786
Query: 140 TLELVS-AGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKI-------PTV 191
+E + A ++I+D A+YLI G +D F +N ++ K+ T+
Sbjct: 787 LVEAAGQSSASSIISD-----AIYLISAGTSD----FVQNYYINPLLNKLYTTDQFSDTL 939
Query: 192 ITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLFNE 251
+ N ++SLY GAR+ V + P+GCLP ++ L + C++S NS A FNE
Sbjct: 940 LRCYSNFIQSLYALGARRIGVTSLPPIGCLPAVITLF---GAHINECVTSLNSDAINFNE 1110
Query: 252 ALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRV 311
L +SQ L+ L LV DIY +DL ++ GF CCG G ++ +
Sbjct: 1111KLNTTSQNLKNMLPGLNLVVFDIYQPLYDLATKPSENGFFEARKACCGTG----LIEVSI 1278
Query: 312 TCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYS 353
C + C S YV WDG H +EAAN +A +++++ S
Sbjct: 1279LCNKKSIGTCANASEYVFWDGFHPSEAANKVLADELITSGIS 1404
>CD409722
Length = 605
Score = 136 bits (343), Expect = 1e-32
Identities = 65/147 (44%), Positives = 92/147 (62%), Gaps = 10/147 (6%)
Frame = -2
Query: 219 GCLPKILAL--AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYA 276
GCLP+ +A LD GC+SS+N AA+ FN L KL+ + D+ + YVDI+
Sbjct: 604 GCLPQNIAKFGTDSSKLDGLGCVSSHNQAAKTFNLQLRALCTKLQGQYPDSNVTYVDIFT 425
Query: 277 IKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQP--------GYQVCDEGSRYV 328
IK LIAN ++YGF P+ CCG+GGPP N+D RV+CG+ + C++ S Y+
Sbjct: 424 IKSSLIANYSRYGFEQPIMACCGYGGPPLNYDSRVSCGETKTFNGTTITAKACNDSSEYI 245
Query: 329 NWDGTHHTEAANTFIASKILSTDYSTP 355
+WDG H+TE AN ++AS+IL+ YS P
Sbjct: 244 SWDGIHYTETANQYVASQILTGKYSDP 164
>TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H11.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(54%)
Length = 855
Score = 127 bits (319), Expect = 8e-30
Identities = 69/197 (35%), Positives = 102/197 (51%)
Frame = +2
Query: 165 DIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKI 224
+IG ND A + ++ + I+K+ I+ + A+++L +GA+ V GCL
Sbjct: 62 EIGVNDYAYTLGSTVSD-ETIRKL--AISSVSGALQTLLEKGAKYLVVQGLPLTGCLTLS 232
Query: 225 LALAQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIAN 284
+ LA D D GC+ S N+ + N L Q+ R + A ++Y D Y ++ N
Sbjct: 233 MYLAPPDDRDDIGCVKSVNNQSYYHNLVLQDKLQEFRKQYPQAVILYADYYDAYRTVMKN 412
Query: 285 ATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIA 344
+K+GF VCCG G PPYNF + TCG P VC S+Y+NWDG H TEA I+
Sbjct: 413 PSKFGFKETFNVCCGSGEPPYNFTVFATCGTPNATVCSSPSQYINWDGVHLTEAMYKVIS 592
Query: 345 SKILSTDYSTPRTPFEF 361
S L +++ P PF F
Sbjct: 593 SMFLQGNFTQP--PFNF 637
>TC203707
Length = 1394
Score = 125 bits (314), Expect = 3e-29
Identities = 107/349 (30%), Positives = 150/349 (42%), Gaps = 18/349 (5%)
Frame = +1
Query: 25 VMMSKCDVPAVLFVFGDSNSDTGG-----LTSGLGFP---INLPNGRTFFHRSTGRLSDG 76
V + + FVFGDS D G T+ P I+ P GR TGR S+G
Sbjct: 79 VRLKGAEAQRAFFVFGDSLVDNGNNNFLATTARADAPPYGIDYPTGRP-----TGRFSNG 243
Query: 77 RLVIDLLCQSLNTRFLTPYLD-SLSGSSFTNGANFAVVGSSTLPKY-VPFSLNIQVMQFL 134
+ D + QSL PYLD L G GANFA G L + F I++ + L
Sbjct: 244 YNIPDFISQSLGAESTLPYLDPELDGERLLVGANFASAGIGILNDTGIQFVNIIRIYRQL 423
Query: 135 HFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSF------AKNLTYVQVIKKI 188
+ + VS + AL LI +G ND +++ A++ Y + +
Sbjct: 424 EYWEEYQQRVSGLIGPEQTERLINGALVLITLGGNDFVNNYYLVPYSARSRQY-NLPDYV 600
Query: 189 PTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARL 248
+I+E + ++ LY GAR+ V TGPLGC+P LA C + AA L
Sbjct: 601 KYIISEYKKVLRRLYEIGARRVLVTGTGPLGCVPAELAQRSTNG----DCSAELQQAAAL 768
Query: 249 FNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFD 308
FN L ++L + + V V+ + D I+N +YGF CCG G PYN
Sbjct: 769 FNPQLVQIIRQLNSEIGSNVFVGVNTQQMHIDFISNPQRYGFVTSKVACCGQG--PYN-- 936
Query: 309 LRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILS--TDYSTP 355
+ P +C Y WD H TE AN I +ILS ++Y P
Sbjct: 937 -GLGLCTPASNLCPNRDSYAFWDPFHPTERANRIIVQQILSGTSEYMYP 1080
>TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment), partial
(35%)
Length = 748
Score = 79.7 bits (195), Expect(2) = 6e-29
Identities = 43/114 (37%), Positives = 63/114 (54%), Gaps = 9/114 (7%)
Frame = +3
Query: 260 LRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQP--- 316
LR L A + YVD+Y +K+ LI++A KYGF + CCG GG YNF+ CG
Sbjct: 255 LRKELPGAAITYVDVYTVKYTLISHAHKYGFEQGVIACCGHGG-KYNFNNTERCGATKRV 431
Query: 317 -GYQV-----CDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFCH 364
G ++ C + S + WDG H+TEAAN +I +I++ +S P + C+
Sbjct: 432 NGTEIVIANSCKDLSVRIIWDGIHYTEAANKWIFQQIVNGSFSDPPHSLKRACY 593
Score = 65.9 bits (159), Expect(2) = 6e-29
Identities = 30/68 (44%), Positives = 39/68 (57%), Gaps = 2/68 (2%)
Frame = +2
Query: 188 IPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKK--DLDLFGCLSSYNSA 245
I V+ + N +K +Y EG FW+HNTGPLGCLP +L K +D FGC +N
Sbjct: 32 IXDVLGQXSNVIKGVYGEGGXSFWIHNTGPLGCLPYMLDRYPMKPTQMDEFGCAKPFNEV 211
Query: 246 ARLFNEAL 253
A+ FN L
Sbjct: 212 AQYFNRKL 235
>TC217084
Length = 1310
Score = 120 bits (301), Expect = 1e-27
Identities = 104/369 (28%), Positives = 159/369 (42%), Gaps = 21/369 (5%)
Frame = +3
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGG------LTSGLGFPINLPN 61
S L L L IT S ++ + + FVFGDS D G +P + +
Sbjct: 87 SSKFLCLCLLITLISI-IVAPQAEAARAFFVFGDSLVDNGNNNFLATTARADSYPYGIDS 263
Query: 62 GRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLD-SLSGSSFTNGANFAVVGSSTLPK 120
HR++GR S+G + DL+ + + + PYL L+G GANFA G L
Sbjct: 264 AS---HRASGRFSNGLNMPDLISEKIGSEPTLPYLSPQLNGERLLVGANFASAGIGILND 434
Query: 121 YVPFSLNI-----QVMQFLHFKARTLELVSAG-AKNVINDEGFRAALYLIDIGQNDLADS 174
+NI Q+ F ++ R L+ +N++N AL LI +G ND ++
Sbjct: 435 TGIQFINIIRITEQLAYFKQYQQRVSALIGEEQTRNLVNK-----ALVLITLGGNDFVNN 599
Query: 175 F------AKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALA 228
+ A++ Y + + +I+E + +LY GAR+ V TGPLGC+P LA+
Sbjct: 600 YYLVPFSARSREYA-LPDYVVFLISEYRKILANLYELGARRVLVTGTGPLGCVPAELAMH 776
Query: 229 QKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKY 288
+ C + A LFN L +L T++ + + + + D ++N Y
Sbjct: 777 SQNG----ECATELQRAVNLFNPQLVQLLHELNTQIGSDVFISANAFTMHLDFVSNPQAY 944
Query: 289 GFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKIL 348
GF CCG G YN + P +C Y WD H +E AN I K +
Sbjct: 945 GFVTSKVACCGQGA--YN---GIGLCTPASNLCPNRDLYAFWDPFHPSERANRLIVDKFM 1109
Query: 349 --STDYSTP 355
ST+Y P
Sbjct: 1110TGSTEYMHP 1136
>TC213374 weakly similar to UP|Q94F40 (Q94F40) At1g28600/F1K23_6, partial
(25%)
Length = 633
Score = 119 bits (297), Expect = 3e-27
Identities = 68/194 (35%), Positives = 94/194 (48%), Gaps = 3/194 (1%)
Frame = +2
Query: 175 FAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQ---KK 231
F + + ++ +P VI I +A+ L GAR V P+GC L + + K
Sbjct: 17 FFQQKSIAEIKSYVPYVINAIASAINELIGLGARTLMVPGNLPIGCSVIYLTIYETIDKT 196
Query: 232 DLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFS 291
FGCL N +N L KLR A ++Y D Y L + TK+GF+
Sbjct: 197 QYGQFGCLKWLNEFGEYYNHKLQSELDKLRVFHPRANIIYADYYNAALPLYRDPTKFGFT 376
Query: 292 NPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTD 351
+ L +CCG GGP YNF+ CG P CD+ S+++ WDG H TEAA FIA ++
Sbjct: 377 D-LKICCGMGGP-YNFNKLTNCGNPSVIACDDPSKHIGWDGVHLTEAAYRFIAKGLIKGP 550
Query: 352 YSTPRTPFEFFCHR 365
YS P+ F C R
Sbjct: 551 YSLPQ--FSTLCFR 586
>BU547342 weakly similar to GP|10764858|gb| F1K23.17 {Arabidopsis thaliana},
partial (6%)
Length = 687
Score = 118 bits (296), Expect = 4e-27
Identities = 67/179 (37%), Positives = 88/179 (48%), Gaps = 3/179 (1%)
Frame = -2
Query: 188 IPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQ---KKDLDLFGCLSSYNS 244
+P VI I +AV L GAR V PLGC L + + K D +GCL N
Sbjct: 632 VPYVIRAITSAVNELIGLGARTLIVPGNLPLGCSINYLTIYETMDKNQYDQYGCLKWLNE 453
Query: 245 AARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPP 304
A +N+ L +LR A ++Y D Y L N T +GF+N L CCG GGP
Sbjct: 452 FAEYYNQKLQSELDRLRGLHSHANIIYADYYNATLPLYHNTTMFGFTN-LKTCCGMGGP- 279
Query: 305 YNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
YN++ CG PG CD+ S+++ WD H TEAA IA ++ Y P+ F FC
Sbjct: 278 YNYNAAADCGDPGAIACDDPSKHIGWDSVHFTEAAYRIIAEGLIKGPYCLPQ--FNTFC 108
>TC218564
Length = 1398
Score = 118 bits (296), Expect = 4e-27
Identities = 104/355 (29%), Positives = 158/355 (44%), Gaps = 20/355 (5%)
Frame = +1
Query: 14 LFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGG---LTSGLGFPINLPNGRTFF-HRS 69
L + +T+S A K +VPAV+ VFGDS+ D+G + + L P GR F R
Sbjct: 46 LLVAVTTSEA-----KNNVPAVI-VFGDSSVDSGNNNVIATVLKSNFK-PYGRDFEGDRP 204
Query: 70 TGRLSDGRLVIDLLCQSLNTRFLTP-YLD-SLSGSSFTNGANFAVVGS------STLPKY 121
TGR +GR+ D + ++ + P YLD + + F G FA G+ S +
Sbjct: 205 TGRFCNGRVPPDFIAEAFGIKRTVPAYLDPAYTIQDFATGVCFASAGTGYDNATSAVLNV 384
Query: 122 VPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTY 181
+P I+ + K RT L A +I++ ALYL+ +G ND +++ Y
Sbjct: 385 IPLWKEIEYYKEYQAKLRT-HLGVEKANKIISE-----ALYLMSLGTNDFLENY-----Y 531
Query: 182 VQVIKKIPTVITEI--------ENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDL 233
V +++ +++ EN V+ LY G RK + GP+GCLP A D
Sbjct: 532 VFPTRRLHFTVSQYQDFLLRIAENFVRELYALGVRKLSITGLGPVGCLPLERATNILGD- 708
Query: 234 DLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNP 293
GC YN A FN L + KL L + + Y+I +D+I + YGF
Sbjct: 709 --HGCNQEYNDVALSFNRKLENVITKLNRELPRLKALSANAYSIVNDIITKPSTYGFEVV 882
Query: 294 LTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKIL 348
CC G F++ C C + +YV WD H TE N ++S ++
Sbjct: 883 EKACCSTG----TFEMSYLCSDKNPLTCTDAEKYVFWDAFHPTEKTNRIVSSYLI 1035
>TC232972 weakly similar to UP|O82681 (O82681) Lanatoside
15'-O-acetylesterase precursor, partial (45%)
Length = 570
Score = 115 bits (289), Expect = 2e-26
Identities = 61/166 (36%), Positives = 87/166 (51%), Gaps = 10/166 (6%)
Frame = +3
Query: 208 RKFWVHNTGPLGCLPKILALA--QKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLK 265
R F V N P+GC P L ++D FGCL SYN+A +N L + ++ R L
Sbjct: 6 RTFMVLNLAPVGCYPAFLVEFPHDSSNIDDFGCLISYNNAVLNYNNMLKETLKQTRESLS 185
Query: 266 DATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQP--------G 317
DA+++YVD +++ +L + T +G CCG+GG YNFD +V+CG
Sbjct: 186 DASVIYVDTHSVLLELFQHPTSHGLQYGTKACCGYGGGDYNFDPKVSCGNTKEINGSIMP 365
Query: 318 YQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
C++ YV+WDG H TEAAN I IL+ +S P F+ C
Sbjct: 366 ATTCNDPYNYVSWDGIHSTEAANKLITFAILNGSFSDPPFIFQEHC 503
>TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase EXL1, partial
(49%)
Length = 1085
Score = 115 bits (287), Expect = 4e-26
Identities = 101/359 (28%), Positives = 164/359 (45%), Gaps = 15/359 (4%)
Frame = +2
Query: 5 LAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGG--LTSGLGFPINLPNG 62
+ +S ++ + L ++S S S +PAV+ VFGDS DTG + + LP G
Sbjct: 44 IPWSFAIVIISLHVSSVSLPNYES---IPAVI-VFGDSIVDTGNNNYITTIAKCNFLPYG 211
Query: 63 RTFF--HRSTGRLSDGRLVIDLLCQSLNTR-FLTPYLDS-------LSGSSFTNGANFAV 112
R F ++ TGR S+G D++ + L PYLD L+G SF +GA+
Sbjct: 212 RDFGGGNQPTGRFSNGLTPSDIIAAKFGVKELLPPYLDPKLQPQDLLTGVSFASGAS--- 382
Query: 113 VGSSTLPKYVPFSLNI--QVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQND 170
G L + +L++ Q+ F +K + +E+V I + ++Y++ G ND
Sbjct: 383 -GYDPLTSKIASALSLSDQLDTFREYKNKIMEIVGENRTATIISK----SIYILCTGSND 547
Query: 171 LADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQK 230
+ +++ + + ++ N ++ LY GAR+ V LGC+P L
Sbjct: 548 ITNTYFVRGGEYDIQAYTDLMASQATNFLQELYGLGARRIGVVGLPVLGCVPSQRTLHG- 724
Query: 231 KDLDLFGCLSSY-NSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYG 289
+F S + N AA LFN L L+ + ++A VY+D+Y +LI N KYG
Sbjct: 725 ---GIFRACSDFENEAAVLFNSKLSSQMDALKKQFQEARFVYLDLYNPVLNLIQNPAKYG 895
Query: 290 FSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKIL 348
F CCG G ++ C +C S Y+ WD H TEAA + +++L
Sbjct: 896 FEVMDQGCCGTG----KLEVGPLCNHFTLLICSNTSNYIFWDSFHPTEAAYNVVCTQVL 1060
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,557,024
Number of Sequences: 63676
Number of extensions: 222618
Number of successful extensions: 1319
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 1170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1178
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0223.2


