

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
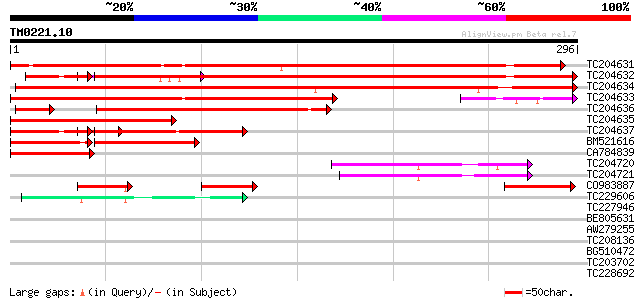
Query= TM0221.10
(296 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204631 weakly similar to UP|NOA1_MOUSE (Q9JKN6) RNA-binding pr... 472 e-133
TC204632 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing prot... 437 e-123
TC204634 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing prot... 399 e-112
TC204633 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {A... 305 2e-83
TC204636 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing prot... 166 6e-46
TC204635 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {A... 162 1e-40
TC204637 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing prot... 145 2e-35
BM521616 93 1e-19
CA784839 76 2e-14
TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 51 5e-07
TC204721 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 47 1e-05
CO983887 45 3e-05
TC229606 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein H... 42 3e-04
TC227946 similar to UP|Q6K6Z4 (Q6K6Z4) KH domain-containing prot... 40 0.001
BE805631 40 0.002
AW279255 39 0.003
TC208136 weakly similar to UP|Q84ZX0 (Q84ZX0) HEN4, partial (13%) 38 0.006
BG510472 36 0.024
TC203702 weakly similar to GB|AAP68215.1|31711718|BT008776 At1g5... 36 0.024
TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, pa... 28 0.050
>TC204631 weakly similar to UP|NOA1_MOUSE (Q9JKN6) RNA-binding protein Nova-1
(Neuro-oncological ventral antigen 1) (Ventral
neuron-specific protein 1) (Fragment), partial (6%)
Length = 1150
Score = 472 bits (1214), Expect = e-133
Identities = 237/294 (80%), Positives = 251/294 (84%), Gaps = 4/294 (1%)
Frame = +2
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MD RKRGRP+ N NG +KK+KQELESLSSGVGSKSKPCTKFFSTAGCPFGE CHFLHY
Sbjct: 128 MDVRKRGRPD--VNLNGAVKKTKQELESLSSGVGSKSKPCTKFFSTAGCPFGEGCHFLHY 301
Query: 61 VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP-AVKTRICNKFNTAEGCKFGDK 119
VPGGYN VAHMMNL P+A PPR VAAPPP +PNGS P AVKTRICNKFNTAEGCKFGDK
Sbjct: 302 VPGGYNVVAHMMNLKPAAP-PPRTVAAPPP-IPNGSAPSAVKTRICNKFNTAEGCKFGDK 475
Query: 120 CHFAHGEWELGKHIAPSFDDH---RPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVE 176
CHFAHGEWELGK IAPS DDH RP+G GR+ GR+EPPP A FGA +TAKISVE
Sbjct: 476 CHFAHGEWELGKPIAPSIDDHHHHRPLGPPAGGRMAGRIEPPPAMAGSFGAISTAKISVE 655
Query: 177 ASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLL 236
ASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEG+F+QIKEASNMVKDLL
Sbjct: 656 ASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGTFEQIKEASNMVKDLL 835
Query: 237 LTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHG 290
LT+ MSAPPKS G G P G +NFKTKLCENFAKG+CTFGERCHFAHG
Sbjct: 836 LTVSMSAPPKSTPGVPGAPAPPG----SNFKTKLCENFAKGSCTFGERCHFAHG 985
Score = 40.0 bits (92), Expect = 0.001
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Frame = +2
Query: 236 LLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENF--AKGTCTFGERCHFAHGPAE 293
++ L+ +APP A P + KT++C F A+G C FG++CHFAHG E
Sbjct: 332 MMNLKPAAPPPRTV--AAPPPIPNGSAPSAVKTRICNKFNTAEG-CKFGDKCHFAHGEWE 502
Query: 294 LRK 296
L K
Sbjct: 503 LGK 511
>TC204632 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing protein-like,
partial (58%)
Length = 1220
Score = 437 bits (1123), Expect = e-123
Identities = 211/253 (83%), Positives = 220/253 (86%), Gaps = 1/253 (0%)
Frame = +2
Query: 45 STAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP-AVKTR 103
STAGCPFGE CHFLHYVPGGYNAVAHMMNL P+A PP A P VPNGS P AVKTR
Sbjct: 275 STAGCPFGEGCHFLHYVPGGYNAVAHMMNLTPAAPPPPSRNVAALPHVPNGSAPSAVKTR 454
Query: 104 ICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPAT 163
ICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHR +G AGR+ GRMEPPPGPA
Sbjct: 455 ICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRAMGPPGAGRLAGRMEPPPGPAA 634
Query: 164 GFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFD 223
FGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSF+
Sbjct: 635 SFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFE 814
Query: 224 QIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGE 283
QIKEASNMVKDLLLTLQMSAPPK+ G G P HG +NFKTKLCENF KG+CTFG+
Sbjct: 815 QIKEASNMVKDLLLTLQMSAPPKTTPGVPGAPASHG----SNFKTKLCENFTKGSCTFGD 982
Query: 284 RCHFAHGPAELRK 296
RCHFAHG +ELRK
Sbjct: 983 RCHFAHGASELRK 1021
Score = 37.0 bits (84), Expect(2) = 2e-06
Identities = 28/76 (36%), Positives = 35/76 (45%), Gaps = 9/76 (11%)
Frame = +2
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPS-----AQAPP--RNVAA- 87
K++ C KF + GC FG+ CHF H G + H +APS A PP +A
Sbjct: 446 KTRICNKFNTAEGCKFGDKCHFAH---GEWELGKH---IAPSFDDHRAMGPPGAGRLAGR 607
Query: 88 -PPPPVPNGSTPAVKT 102
PPP P S A T
Sbjct: 608 MEPPPGPAASFGANAT 655
Score = 31.6 bits (70), Expect(2) = 2e-06
Identities = 16/35 (45%), Positives = 22/35 (62%)
Frame = +3
Query: 9 PEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKF 43
P+PGF+ NGG K+SKQ L L + + + CT F
Sbjct: 228 PKPGFSLNGGFKRSKQAL--LVAHLVRAATSCTMF 326
Score = 30.0 bits (66), Expect = 1.3
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = +2
Query: 30 SSGVGSKSKPCTKFFSTAGCPFGESCHFLH 59
S G K+K C F + C FG+ CHF H
Sbjct: 914 SHGSNFKTKLCENF-TKGSCTFGDRCHFAH 1000
>TC204634 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing protein-like,
partial (67%)
Length = 1248
Score = 399 bits (1025), Expect = e-112
Identities = 194/299 (64%), Positives = 231/299 (76%), Gaps = 6/299 (2%)
Frame = +1
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
RKRGRPE FN NGG KKS+ E +S +G+GSKSKPCTKFFST+GCPFGE CHFLHYVPG
Sbjct: 205 RKRGRPEAAFNFNGGAKKSRPETDSFPTGLGSKSKPCTKFFSTSGCPFGEGCHFLHYVPG 384
Query: 64 GYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGST-PAVKTRICNKFNTAEGCKFGDKCHF 122
G+ AV+ ++N+ + P PP P+GS+ P VKTR+CNKFNTAEGCKFGDKCHF
Sbjct: 385 GFKAVSQLINVGSNPVTPQVGRNPVPPSFPDGSSPPVVKTRLCNKFNTAEGCKFGDKCHF 564
Query: 123 AHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPP---PGPATGFGANATAKISVEASL 179
AHGEWELG+ APS++D R +G P+ R+GGR+EPP G A FGA+ATAKIS+ ASL
Sbjct: 565 AHGEWELGRPTAPSYEDPRVMGQMPSSRVGGRVEPPHPAHGAAASFGASATAKISINASL 744
Query: 180 AGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLTL 239
AGA+IGK GVNSKQICR TGAKLSIR+H++DPNLRNIELEGSFDQIK+AS MV +++L +
Sbjct: 745 AGAVIGKNGVNSKQICRVTGAKLSIRDHDTDPNLRNIELEGSFDQIKQASAMVHEVILNV 924
Query: 240 QMSA--PPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
++ P KS P ++NFKTKLCENFAKG+CTFGERCHFAHG ELRK
Sbjct: 925 SSASGPPMKSFTSQTSAP-------TSNFKTKLCENFAKGSCTFGERCHFAHGTDELRK 1080
>TC204633 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {Arabidopsis
thaliana;} , partial (38%)
Length = 741
Score = 305 bits (780), Expect = 2e-83
Identities = 145/172 (84%), Positives = 151/172 (87%), Gaps = 1/172 (0%)
Frame = +3
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MD RKRGRPEPGF+ NGG KKSKQE+ESLS+GVGSKSKPCTKFFSTAGCPFGE CHFLHY
Sbjct: 228 MDTRKRGRPEPGFSLNGGFKKSKQEMESLSTGVGSKSKPCTKFFSTAGCPFGEGCHFLHY 407
Query: 61 VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP-AVKTRICNKFNTAEGCKFGDK 119
VPGGYNAVAHMMNL P+A RNVAA P VPNGS P AVKTRICNKFNTAEGCKFGDK
Sbjct: 408 VPGGYNAVAHMMNLTPAAPPASRNVAALPH-VPNGSAPSAVKTRICNKFNTAEGCKFGDK 584
Query: 120 CHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATA 171
CHFAHGEWELGKHIAPSFDDHR +G AGR+ GRMEPPPGPA FGANATA
Sbjct: 585 CHFAHGEWELGKHIAPSFDDHRAMGPPGAGRLAGRMEPPPGPAASFGANATA 740
Score = 41.2 bits (95), Expect = 6e-04
Identities = 27/65 (41%), Positives = 37/65 (56%), Gaps = 4/65 (6%)
Frame = +3
Query: 236 LLTLQMSAPPKSNQGGAGGPGGHGHHGS--NNFKTKLCENF--AKGTCTFGERCHFAHGP 291
++ L +APP S A H +GS + KT++C F A+G C FG++CHFAHG
Sbjct: 438 MMNLTPAAPPASRNVAALP---HVPNGSAPSAVKTRICNKFNTAEG-CKFGDKCHFAHGE 605
Query: 292 AELRK 296
EL K
Sbjct: 606 WELGK 620
Score = 28.1 bits (61), Expect = 4.9
Identities = 12/24 (50%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = +3
Query: 267 KTKLCENF-AKGTCTFGERCHFAH 289
K+K C F + C FGE CHF H
Sbjct: 333 KSKPCTKFFSTAGCPFGEGCHFLH 404
>TC204636 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing protein-like,
partial (21%)
Length = 654
Score = 166 bits (421), Expect(2) = 6e-46
Identities = 75/124 (60%), Positives = 93/124 (74%), Gaps = 1/124 (0%)
Frame = +1
Query: 46 TAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP-AVKTRI 104
T+GCPFGE CHFLHYVPGG+ AV+ ++N+ + P PP P+GS+P VKTR+
Sbjct: 286 TSGCPFGEGCHFLHYVPGGFKAVSQLINVGSNPVTPQVGRNPVPPSFPDGSSPPVVKTRL 465
Query: 105 CNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATG 164
CNKFNTAEGCKFGDKCHFAHGEWELG+ APS++D R +G P+ R+GGR+E PP PA G
Sbjct: 466 CNKFNTAEGCKFGDKCHFAHGEWELGRPTAPSYEDPRVMGQMPSSRVGGRVE-PPHPAHG 642
Query: 165 FGAN 168
A+
Sbjct: 643 AAAS 654
Score = 38.1 bits (87), Expect = 0.005
Identities = 18/30 (60%), Positives = 22/30 (73%), Gaps = 2/30 (6%)
Frame = +1
Query: 267 KTKLCENF--AKGTCTFGERCHFAHGPAEL 294
KT+LC F A+G C FG++CHFAHG EL
Sbjct: 454 KTRLCNKFNTAEG-CKFGDKCHFAHGEWEL 540
Score = 35.4 bits (80), Expect = 0.031
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +1
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH 59
K++ C KF + GC FG+ CHF H
Sbjct: 454 KTRLCNKFNTAEGCKFGDKCHFAH 525
Score = 35.4 bits (80), Expect(2) = 6e-46
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +2
Query: 4 RKRGRPEPGFNSNGGIKKSK 23
RKRGRPE FN NGG KKS+
Sbjct: 221 RKRGRPEAAFNFNGGAKKSR 280
>TC204635 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {Arabidopsis
thaliana;} , partial (30%)
Length = 464
Score = 162 bits (411), Expect = 1e-40
Identities = 76/88 (86%), Positives = 80/88 (90%), Gaps = 1/88 (1%)
Frame = +3
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MD RKRGRPEPGF+ NGG KKSKQE+ESLS+GVGSKSKPCTKFFSTAGCPFGE CHFLHY
Sbjct: 198 MDIRKRGRPEPGFSLNGGFKKSKQEMESLSTGVGSKSKPCTKFFSTAGCPFGEGCHFLHY 377
Query: 61 VPGGYNAVAHMMNLAPSAQAPP-RNVAA 87
VPGGYNAVAHMMNL P+A PP RNVAA
Sbjct: 378 VPGGYNAVAHMMNLTPAAPLPPTRNVAA 461
Score = 34.7 bits (78), Expect = 0.053
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +3
Query: 101 KTRICNKFNTAEGCKFGDKCHFAH 124
K++ C KF + GC FG+ CHF H
Sbjct: 303 KSKPCTKFFSTAGCPFGEGCHFLH 374
Score = 28.1 bits (61), Expect = 4.9
Identities = 12/24 (50%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = +3
Query: 267 KTKLCENF-AKGTCTFGERCHFAH 289
K+K C F + C FGE CHF H
Sbjct: 303 KSKPCTKFFSTAGCPFGEGCHFLH 374
>TC204637 similar to UP|Q7F8R0 (Q7F8R0) KH domain-containing protein-like,
partial (21%)
Length = 503
Score = 145 bits (366), Expect = 2e-35
Identities = 69/81 (85%), Positives = 70/81 (86%), Gaps = 1/81 (1%)
Frame = +3
Query: 45 STAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGST-PAVKTR 103
STAGCPFGE CHFLHYVPGGYNAVAHMMNL P+A RNVAA PPVPNGS PAVKTR
Sbjct: 264 STAGCPFGEGCHFLHYVPGGYNAVAHMMNLTPAAPPASRNVAA-LPPVPNGSAPPAVKTR 440
Query: 104 ICNKFNTAEGCKFGDKCHFAH 124
ICNKFNTAEGCK GDKCHFAH
Sbjct: 441 ICNKFNTAEGCKXGDKCHFAH 503
Score = 47.8 bits (112), Expect(2) = 8e-10
Identities = 25/43 (58%), Positives = 29/43 (67%)
Frame = +1
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKF 43
MD RKRGRPEPGF+ NGG KKSKQ L L + + + CT F
Sbjct: 193 MDTRKRGRPEPGFSLNGGFKKSKQAL--LVAHLVRAATSCTMF 315
Score = 35.0 bits (79), Expect = 0.040
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Frame = +3
Query: 236 LLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENF--AKGTCTFGERCHFAH 289
++ L +APP S A P +G KT++C F A+G C G++CHFAH
Sbjct: 342 MMNLTPAAPPASRNVAALPPVPNGS-APPAVKTRICNKFNTAEG-CKXGDKCHFAH 503
Score = 32.7 bits (73), Expect(2) = 8e-10
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = +3
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH 59
K++ C KF + GC G+ CHF H
Sbjct: 432 KTRICNKFNTAEGCKXGDKCHFAH 503
>BM521616
Length = 428
Score = 93.2 bits (230), Expect = 1e-19
Identities = 41/55 (74%), Positives = 43/55 (77%)
Frame = +3
Query: 45 STAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
STAGCPFGE CHFLHYVPGGYNAVAHMMNL P+A PP A P VPNGS P+
Sbjct: 255 STAGCPFGEGCHFLHYVPGGYNAVAHMMNLTPAAPLPPTRNVAALPHVPNGSAPS 419
Score = 48.1 bits (113), Expect = 5e-06
Identities = 26/43 (60%), Positives = 28/43 (64%)
Frame = +1
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKF 43
MD RKRGRPEPGF+ NGG KKSKQ L S + S CT F
Sbjct: 184 MDIRKRGRPEPGFSLNGGFKKSKQALLVAHSVRAATS--CTMF 306
>CA784839
Length = 438
Score = 76.3 bits (186), Expect = 2e-14
Identities = 35/44 (79%), Positives = 39/44 (88%)
Frame = +1
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFF 44
MD RKRGR E GF+ NGG K+SKQE+ESLS+GVGSKSKPCTKFF
Sbjct: 115 MDIRKRGRHETGFSLNGGFKRSKQEMESLSTGVGSKSKPCTKFF 246
>TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (31%)
Length = 1936
Score = 51.2 bits (121), Expect = 5e-07
Identities = 36/110 (32%), Positives = 57/110 (51%), Gaps = 5/110 (4%)
Frame = +3
Query: 169 ATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQI 225
A+ KI + G IIGKGG K + Q+GAK+ + R+ ++DPN R +EL GS D I
Sbjct: 120 ASKKIDIPNGRVGVIIGKGGETIKYLQLQSGAKIQVTRDMDADPNSATRTVELMGSPDAI 299
Query: 226 KEASNMVKDLLLTLQMSAPPKSNQGGAG--GPGGHGHHGSNNFKTKLCEN 273
A ++ ++L ++ GG+G G GS+ + +K+ N
Sbjct: 300 ATAEKLINEVL--------AEAETGGSGIIARRVAGQAGSDEYVSKIPNN 425
Score = 35.4 bits (80), Expect = 0.031
Identities = 19/60 (31%), Positives = 32/60 (52%), Gaps = 4/60 (6%)
Frame = +3
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDP----NLRNIELEGSFDQIKEASNMVKDLL 236
G +IGKGG K + TGA++ + P R +++EG+ +QI+ A MV ++
Sbjct: 432 GLVIGKGGETIKNMQASTGARIQVIPLHLPPGDTSTERTLKIEGTPEQIESAKQMVNQVI 611
>TC204721 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (30%)
Length = 1762
Score = 46.6 bits (109), Expect = 1e-05
Identities = 34/104 (32%), Positives = 51/104 (48%), Gaps = 3/104 (2%)
Frame = +2
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQIKEAS 229
I + G IIGKGG K + Q+GAK+ I R+ ++DPN R +EL G+ + I A
Sbjct: 2 IDIPNGRVGVIIGKGGETIKYLQLQSGAKIQITRDIDADPNSSTRTVELMGTPEAISSAE 181
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCEN 273
++ ++L A +S G G GS+ F K+ N
Sbjct: 182 KLINEVL------AEAESGGSGIVTRRFTGQAGSDEFVMKIPNN 295
Score = 33.5 bits (75), Expect = 0.12
Identities = 20/78 (25%), Positives = 38/78 (48%), Gaps = 4/78 (5%)
Frame = +2
Query: 163 TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP----NLRNIEL 218
TG + + + + G IIGKGG K + TGA++ + P R +++
Sbjct: 248 TGQAGSDEFVMKIPNNKVGLIIGKGGETIKNMQASTGARIQVIPLHLPPGDTSTERTLKI 427
Query: 219 EGSFDQIKEASNMVKDLL 236
+G+ +QI+ A +V ++
Sbjct: 428 DGTPEQIESAKQLVYQVI 481
>CO983887
Length = 795
Score = 45.4 bits (106), Expect = 3e-05
Identities = 18/29 (62%), Positives = 21/29 (72%)
Frame = -2
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT++C KF T C FGD CHFAHG+ EL
Sbjct: 638 KTKLCIKFETTGHCPFGDDCHFAHGQAEL 552
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/38 (55%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Frame = -2
Query: 259 GHHGSNNFKTKLCENF-AKGTCTFGERCHFAHGPAELR 295
G+ S +KTKLC F G C FG+ CHFAHG AEL+
Sbjct: 662 GNVKSTYWKTKLCIKFETTGHCPFGDDCHFAHGQAELQ 549
Score = 40.8 bits (94), Expect = 7e-04
Identities = 18/34 (52%), Positives = 21/34 (60%), Gaps = 5/34 (14%)
Frame = -2
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH-----YVPGG 64
K+K C KF +T CPFG+ CHF H VPGG
Sbjct: 638 KTKLCIKFETTGHCPFGDDCHFAHGQAELQVPGG 537
>TC229606 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein HUA1, partial
(50%)
Length = 1569
Score = 42.4 bits (98), Expect = 3e-04
Identities = 34/122 (27%), Positives = 48/122 (38%), Gaps = 4/122 (3%)
Frame = +3
Query: 7 GRPEPGFNSNGGIKKSKQELESLSSGVGSK--SKPCTKFFSTAGCPFGESCHFLH--YVP 62
G+ E +++N K+ + E S + + K C + T C FG+SC F H +VP
Sbjct: 168 GQSEAWYSTNPLAKRPRYESASNMTIYPQRPGEKDCAHYMLTRTCKFGDSCKFDHPFWVP 347
Query: 63 GGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHF 122
G P + P + PP P C F + CKFG KC F
Sbjct: 348 EGG---------IPDWKEVPIVTSETPPERPGEPD-------CPYFLKTQRCKFGSKCKF 479
Query: 123 AH 124
H
Sbjct: 480 NH 485
Score = 32.3 bits (72), Expect = 0.26
Identities = 22/85 (25%), Positives = 27/85 (30%)
Frame = +3
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C + T C FGE C + H + AP + A P G
Sbjct: 1041 CDFYMKTGECKFGERCKYHHPI---------------DRSAPSLSKQATVKLTPAGLPRR 1175
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAH 124
IC + CKFG C F H
Sbjct: 1176 EGAVICPYYLKTGTCKFGATCKFDH 1250
Score = 30.0 bits (66), Expect = 1.3
Identities = 30/136 (22%), Positives = 46/136 (33%), Gaps = 14/136 (10%)
Frame = +3
Query: 3 FRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSK--PCTKFFSTAGCPFGESCHFLH- 59
F K R + G K E +SSG+ + PC + T C +G +C F H
Sbjct: 435 FLKTQRCKFGSKCKFNHPKVSSENADVSSGLPERPSEPPCAFYMKTGKCRYGAACKFHHP 614
Query: 60 ------YVPGGYNAVAH-----MMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKF 108
VA +M A P +++ +P G + C +
Sbjct: 615 KDIQIQLSNDSSQTVAQTQTNSIMGWATGDTPPIQSLISPSLQNSKGLPVRLGEVDCPFY 794
Query: 109 NTAEGCKFGDKCHFAH 124
CK+G C + H
Sbjct: 795 MKTGSCKYGVTCRYNH 842
Score = 28.5 bits (62), Expect = 3.8
Identities = 10/23 (43%), Positives = 11/23 (47%)
Frame = +3
Query: 105 CNKFNTAEGCKFGDKCHFAHGEW 127
C + CKFGD C F H W
Sbjct: 273 CAHYMLTRTCKFGDSCKFDHPFW 341
Score = 27.7 bits (60), Expect = 6.5
Identities = 20/85 (23%), Positives = 25/85 (28%)
Frame = +3
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C F T C FG C F H N A + P + PP
Sbjct: 426 CPYFLKTQRCKFGSKCKFNHPKVSSEN--------ADVSSGLPERPSEPP---------- 551
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAH 124
C + C++G C F H
Sbjct: 552 -----CAFYMKTGKCRYGAACKFHH 611
>TC227946 similar to UP|Q6K6Z4 (Q6K6Z4) KH domain-containing protein
NOVA-like, partial (58%)
Length = 1228
Score = 40.0 bits (92), Expect = 0.001
Identities = 36/118 (30%), Positives = 53/118 (44%), Gaps = 15/118 (12%)
Frame = +2
Query: 157 PPPGPATGFGANAT-AKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNL- 213
PP P+ T + V S AG++IGKGG Q+GA++ + R HE P
Sbjct: 476 PPKSPSLDSEEKPTYIRFLVSNSAAGSVIGKGGSTITDFQSQSGARIQLSRNHEFFPGTT 655
Query: 214 -RNIELEGSFDQIKEASNMVKDLLLTLQMS-----APPKSN------QGGAGGPGGHG 259
R I + G+ ++I+ A ++ LL+ S A PK+ G GG G G
Sbjct: 656 DRIIMVSGAINEIQRAVELILSKLLSELHSEDDNDAEPKTKVRLVVPNGSCGGIIGKG 829
Score = 30.4 bits (67), Expect = 1.00
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Frame = +2
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESD---PNLRNIELEGSFDQIKEA 228
++ V G IIGKGG + + A + I +++ N R + L GSFD+ A
Sbjct: 782 RLVVPNGSCGGIIGKGGATIRSFIEDSQAGIKISPQDNNYYGQNDRLVTLTGSFDEQMRA 961
Query: 229 SNMV 232
++
Sbjct: 962 IELI 973
>BE805631
Length = 196
Score = 39.7 bits (91), Expect = 0.002
Identities = 16/29 (55%), Positives = 18/29 (61%)
Frame = +2
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT +C + CKFGDKC FAHG EL
Sbjct: 5 KTEMCKNWELRGACKFGDKCCFAHGRHEL 91
Score = 38.9 bits (89), Expect = 0.003
Identities = 15/31 (48%), Positives = 24/31 (77%), Gaps = 1/31 (3%)
Frame = +2
Query: 266 FKTKLCENFA-KGTCTFGERCHFAHGPAELR 295
+KT++C+N+ +G C FG++C FAHG EL+
Sbjct: 2 YKTEMCKNWELRGACKFGDKCCFAHGRHELK 94
Score = 27.3 bits (59), Expect = 8.4
Identities = 9/24 (37%), Positives = 15/24 (62%)
Frame = +2
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH 59
K+K C ++ + CP+G C +LH
Sbjct: 122 KTKRC*QYHHSGYCPYGLRCQYLH 193
>AW279255
Length = 409
Score = 38.9 bits (89), Expect = 0.003
Identities = 29/99 (29%), Positives = 48/99 (48%), Gaps = 15/99 (15%)
Frame = +1
Query: 181 GAIIGKGGVNSKQICRQTGAKLS-IREH---ESDPNLRNIELEGSFDQIKEASNMVKDLL 236
G IIG+GG K + ++GA++ I +H D R +++ G QI+ A ++K++
Sbjct: 25 GLIIGRGGETIKSLQTKSGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEIAQELIKEV- 201
Query: 237 LTLQMSAPPKSNQG-----------GAGGPGGHGHHGSN 264
M+ P K + G G+GGP G GS+
Sbjct: 202 ----MNQPVKPSSGGFGQQAYRPPRGSGGPPQWGQRGSH 306
>TC208136 weakly similar to UP|Q84ZX0 (Q84ZX0) HEN4, partial (13%)
Length = 1163
Score = 37.7 bits (86), Expect = 0.006
Identities = 25/92 (27%), Positives = 47/92 (50%), Gaps = 10/92 (10%)
Frame = +1
Query: 166 GANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP-----NLRNIELEG 220
G+ T ++ V +S G +IGKGGV ++ + TGA + I + P N + +++ G
Sbjct: 334 GSIVTVRLVVPSSQVGCLIGKGGVIVSEMRKATGANIRIIGTDQVPKCASDNDQVVQISG 513
Query: 221 SFDQIKEA-----SNMVKDLLLTLQMSAPPKS 247
F +++A + +L ++ Q SA +S
Sbjct: 514 EFSSVQDALYNAMGRLRDNLFVSTQNSAGTRS 609
>BG510472
Length = 428
Score = 35.8 bits (81), Expect = 0.024
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +1
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKTKLC + +G C C FAHG A+LR+
Sbjct: 241 FKTKLCVLYQRGRCN-RHNCSFAHGSADLRR 330
>TC203702 weakly similar to GB|AAP68215.1|31711718|BT008776 At1g51580
{Arabidopsis thaliana;} , partial (25%)
Length = 1642
Score = 35.8 bits (81), Expect = 0.024
Identities = 24/91 (26%), Positives = 45/91 (49%), Gaps = 12/91 (13%)
Frame = +3
Query: 167 ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHES----DPNLRNIELEGSF 222
A AK+ V + G ++GKGG+ ++ R TGA + I E N +++ GS
Sbjct: 150 AAVVAKLLVRSPQVGCLLGKGGLVISEMRRVTGASIRIFSKEQIKYISQNEEVVQVIGSL 329
Query: 223 DQIKEA----SNMVKDLLLTLQ----MSAPP 245
+++A +N +++ + ++ SAPP
Sbjct: 330 QSVQDALFHITNRIRETIFPIRTPPNFSAPP 422
>TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, partial (48%)
Length = 775
Score = 28.5 bits (62), Expect(2) = 0.050
Identities = 16/43 (37%), Positives = 18/43 (41%)
Frame = +2
Query: 119 KCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGP 161
+CH H L H + HRP H P R R PPP P
Sbjct: 272 RCHRRHPLHHLPPHPS-DLHPHRPPSHGPHLRALPRHRPPPPP 397
Score = 28.1 bits (61), Expect = 4.9
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 2/29 (6%)
Frame = +1
Query: 76 PSAQAPPRNVA--APPPPVPNGSTPAVKT 102
PS PP N + +PPPP P+ ST ++++
Sbjct: 238 PSPATPPPNTSPLSPPPPPPSPSTASLRS 324
Score = 25.0 bits (53), Expect(2) = 0.050
Identities = 10/22 (45%), Positives = 12/22 (54%)
Frame = +1
Query: 81 PPRNVAAPPPPVPNGSTPAVKT 102
PP PPPP P+ +TP T
Sbjct: 202 PPXPRHXPPPPPPSPATPPPNT 267
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,806,785
Number of Sequences: 63676
Number of extensions: 300831
Number of successful extensions: 4317
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 3016
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4089
length of query: 296
length of database: 12,639,632
effective HSP length: 97
effective length of query: 199
effective length of database: 6,463,060
effective search space: 1286148940
effective search space used: 1286148940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0221.10


