

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.25
(334 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
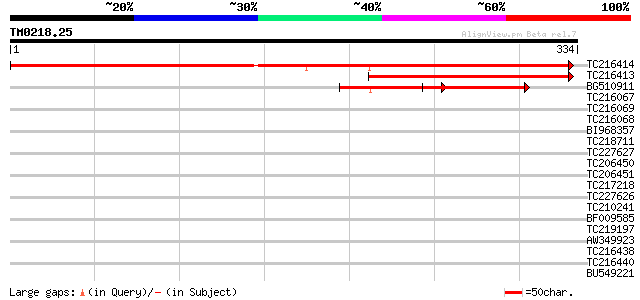
TC216414 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, part... 631 0.0
TC216413 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, part... 220 8e-58
BG510911 118 4e-27
TC216067 weakly similar to UP|RH2A_ARATH (Q9ZT50) RHA2a protein ... 39 0.004
TC216069 weakly similar to UP|RH2A_ARATH (Q9ZT50) RHA2a protein ... 37 0.010
TC216068 similar to UP|Q9ZU51 (Q9ZU51) RING-H2 finger protein RH... 37 0.016
BI968357 36 0.021
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 33 0.14
TC227627 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (49%) 33 0.18
TC206450 33 0.24
TC206451 33 0.24
TC217218 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MB... 32 0.31
TC227626 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (34%) 32 0.40
TC210241 weakly similar to UP|Q9SD55 (Q9SD55) Zinc-finger-like p... 32 0.40
BF009585 similar to GP|2970051|dbj ARG10 {Vigna radiata}, partia... 32 0.53
TC219197 31 0.69
AW349923 weakly similar to PIR|T00747|T007 RING-H2 finger protei... 31 0.90
TC216438 homologue to UP|Q8GUU2 (Q8GUU2) RES protein, partial (14%) 31 0.90
TC216440 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (68%) 31 0.90
BU549221 30 2.0
>TC216414 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, partial (88%)
Length = 1565
Score = 631 bits (1627), Expect = 0.0
Identities = 303/339 (89%), Positives = 318/339 (93%), Gaps = 7/339 (2%)
Frame = +3
Query: 1 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQAV 60
MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICV+RTYSEWVIDGEYDWPPKCCQCQ+V
Sbjct: 207 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVVRTYSEWVIDGEYDWPPKCCQCQSV 386
Query: 61 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSR 120
LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGY CP+CST+IWPPKSVKDSGSR
Sbjct: 387 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYVCPSCSTTIWPPKSVKDSGSR 566
Query: 121 LHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD-----SP 175
LHSKLKEAIMQ+GMEKN+FGNHPV SVTESRSPPPAFAS+PLI RENHGNSD SP
Sbjct: 567 LHSKLKEAIMQSGMEKNVFGNHPV--SVTESRSPPPAFASDPLIIRENHGNSDSVDGYSP 740
Query: 176 ATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVGP--ATRKGAFNVERQNSEISYYADDE 233
ATGSE PKLSVTDIVEI+GAN AGNF+K SSPVGP TRKG+ +VERQNSEISYYADDE
Sbjct: 741 ATGSELPKLSVTDIVEIDGANPAGNFMKNSSPVGPGATTRKGSVHVERQNSEISYYADDE 920
Query: 234 DGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSR 293
D NRKKYTKRGPF+HKFLRALLPFWS+ALPTLPVTAPPRKDASNA E SEGRTRHQRSSR
Sbjct: 921 DANRKKYTKRGPFHHKFLRALLPFWSTALPTLPVTAPPRKDASNAAETSEGRTRHQRSSR 1100
Query: 294 MDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPNEE 332
MDPRKILLLIAIMACMATMGILYYRL QRGPG+EL N+E
Sbjct: 1101MDPRKILLLIAIMACMATMGILYYRLAQRGPGDELRNDE 1217
>TC216413 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, partial (32%)
Length = 519
Score = 220 bits (560), Expect = 8e-58
Identities = 109/122 (89%), Positives = 114/122 (93%), Gaps = 1/122 (0%)
Frame = +3
Query: 212 TRKGAFNVERQNSEISYYADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPP 271
TRKG +VERQNSEISYYADDED NRKKYTKRGPF+HKFLRALLPFWS+ALPTLPVTAPP
Sbjct: 12 TRKGPVHVERQNSEISYYADDEDANRKKYTKRGPFHHKFLRALLPFWSTALPTLPVTAPP 191
Query: 272 RKDASN-ATEGSEGRTRHQRSSRMDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPN 330
RKDASN A E SEGRTRHQRSSRMDPRKILLLIAIMACMATMGILYYRL QRGPG+EL N
Sbjct: 192 RKDASNAAAETSEGRTRHQRSSRMDPRKILLLIAIMACMATMGILYYRLAQRGPGDELLN 371
Query: 331 EE 332
+E
Sbjct: 372 DE 377
>BG510911
Length = 440
Score = 118 bits (295), Expect = 4e-27
Identities = 59/64 (92%), Positives = 61/64 (95%), Gaps = 1/64 (1%)
Frame = +3
Query: 244 GPFYHKFLRALLPFWSSALPTLPVTAPPRKDASN-ATEGSEGRTRHQRSSRMDPRKILLL 302
GPF+HKFLRALLPFWS+ALPTLPVTAPPRKDASN A E SEGRTRHQRSSRMDPRKILLL
Sbjct: 249 GPFHHKFLRALLPFWSTALPTLPVTAPPRKDASNAAAETSEGRTRHQRSSRMDPRKILLL 428
Query: 303 IAIM 306
IAIM
Sbjct: 429 IAIM 440
Score = 85.5 bits (210), Expect = 3e-17
Identities = 46/65 (70%), Positives = 48/65 (73%), Gaps = 2/65 (3%)
Frame = +1
Query: 195 ANSAGNFVKGSSPVGPA--TRKGAFNVERQNSEISYYADDEDGNRKKYTKRGPFYHKFLR 252
AN AGNF+K SSPVGP TRKG +VERQNSEISYYADDED NRKKYTKRG FL
Sbjct: 1 ANPAGNFMKNSSPVGPGATTRKGPVHVERQNSEISYYADDEDANRKKYTKRGDIV--FLN 174
Query: 253 ALLPF 257
L F
Sbjct: 175 GFLIF 189
>TC216067 weakly similar to UP|RH2A_ARATH (Q9ZT50) RHA2a protein (RING-H2
zinc finger protein), partial (37%)
Length = 733
Score = 38.5 bits (88), Expect = 0.004
Identities = 24/81 (29%), Positives = 36/81 (43%)
Frame = +3
Query: 45 DGEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTC 104
D ++D P C CQA E+GD Q L C HV H C + + + CP C
Sbjct: 342 DHDHDHHP-CVVCQATFEDGD--QVRMLPCRHVFHRRCFDGWLHHYK-------FNCPLC 491
Query: 105 STSIWPPKSVKDSGSRLHSKL 125
+ ++ + V + RL +L
Sbjct: 492 RSPLFSDERVALTERRLGQQL 554
>TC216069 weakly similar to UP|RH2A_ARATH (Q9ZT50) RHA2a protein (RING-H2
zinc finger protein), partial (37%)
Length = 497
Score = 37.4 bits (85), Expect = 0.010
Identities = 24/80 (30%), Positives = 35/80 (43%)
Frame = +1
Query: 46 GEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCS 105
G +D P C CQA E+GD Q L C HV H C + + + CP C
Sbjct: 31 GGHDHHP-CVVCQAXXEDGD--QVRMLPCRHVFHRRCFDGWLHHYK-------FNCPLCR 180
Query: 106 TSIWPPKSVKDSGSRLHSKL 125
+ ++ + V + RL +L
Sbjct: 181 SPLFSDERVAVTERRLGQQL 240
>TC216068 similar to UP|Q9ZU51 (Q9ZU51) RING-H2 finger protein RHA2b, partial
(22%)
Length = 1060
Score = 36.6 bits (83), Expect = 0.016
Identities = 21/72 (29%), Positives = 31/72 (42%)
Frame = +3
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKS 113
C CQA E+GD Q L C HV H C + + + CP C + ++ +
Sbjct: 393 CVVCQATFEDGD--QVRMLPCRHVFHRRCFDGWLHHYK-------FNCPLCRSPLFSDER 545
Query: 114 VKDSGSRLHSKL 125
V + RL +L
Sbjct: 546 VALTERRLGQQL 581
>BI968357
Length = 396
Score = 36.2 bits (82), Expect = 0.021
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 2/101 (1%)
Frame = -1
Query: 23 GECICFPEHQICVIR-TYSEWVIDGEYDWPPKCCQCQAVLEE-GDGSQTTRLGCLHVIHT 80
G C+C +CV + S W++ G + C LE+ G Q L C H H+
Sbjct: 384 GVCVC-----VCVCSLSLSLWLLRGN-KFKKDRKVCAVCLEDLGQEQQVMNLSCSHKYHS 223
Query: 81 NCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRL 121
CLV + + P CP C ++ P ++ GSRL
Sbjct: 222 ACLVPWVAAHP--------HCPYCRAAVQPWPDIQ--GSRL 130
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 33.5 bits (75), Expect = 0.14
Identities = 28/101 (27%), Positives = 44/101 (42%), Gaps = 6/101 (5%)
Frame = -3
Query: 91 PPHTAPAGYACPTCSTSIWP----PKSVKDSGSRLHSKLKE--AIMQTGMEKNIFGNHPV 144
PP T +A PT +TSI P P D G ++ K+++ A++ G E + + P
Sbjct: 1055 PPETLVQKFAYPTVTTSIDPTIPVPAPPIDQGQKMRKKMRKRSALISKGAEPDEHDDKPP 876
Query: 145 SLSVTESRSPPPAFASEPLIGRENHGNSDSPATGSEPPKLS 185
V S P + +P I D PAT +P ++
Sbjct: 875 GTPVQRSIHPIAEPSIDPTIPASQTPPVD-PATPVDPEPMT 756
>TC227627 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (49%)
Length = 967
Score = 33.1 bits (74), Expect = 0.18
Identities = 21/80 (26%), Positives = 32/80 (39%)
Frame = +3
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPK 112
+CC C E DG++ L C H H++C+V +K CP C +I
Sbjct: 486 ECCICLCSYE--DGAELHALPCNHHFHSSCIVKWLK--------MNATCPLCKYNILKGN 635
Query: 113 SVKDSGSRLHSKLKEAIMQT 132
S H+K + + T
Sbjct: 636 EQV*QKSEEHTKYQLGVSTT 695
>TC206450
Length = 752
Score = 32.7 bits (73), Expect = 0.24
Identities = 19/60 (31%), Positives = 25/60 (41%), Gaps = 1/60 (1%)
Frame = +1
Query: 57 CQAVLEE-GDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVK 115
C LE+ G Q L C H H+ CL+ + S P CP C T + P +K
Sbjct: 469 CAVCLEDLGLEQQVMNLSCSHKYHSACLLRWLASHP--------HCPYCRTPVQPSPHIK 624
>TC206451
Length = 1035
Score = 32.7 bits (73), Expect = 0.24
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Frame = +2
Query: 57 CQAVLEE-GDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVK 115
C LE+ G Q L C H H+ CL+ + + P CP C T + P ++
Sbjct: 428 CAVCLEDLGQEQQVMNLSCSHKYHSACLLPWLAAHP--------HCPYCRTPVQP*PHIQ 583
Query: 116 DSGSRL 121
GSRL
Sbjct: 584 --GSRL 595
>TC217218 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MBD2_14
{Arabidopsis thaliana;} , partial (14%)
Length = 1087
Score = 32.3 bits (72), Expect = 0.31
Identities = 19/54 (35%), Positives = 25/54 (46%)
Frame = +3
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTS 107
CC CQ + GDG+ L C H H++C IK + H CP C T+
Sbjct: 546 CCVCQE--DYGDGNDIGTLDCGHDFHSSC----IKQWLMHK----NLCPICKTT 677
>TC227626 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (34%)
Length = 834
Score = 32.0 bits (71), Expect = 0.40
Identities = 17/56 (30%), Positives = 25/56 (44%)
Frame = +2
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
+CC C E DG++ L C H H++C+V +K CP C +I
Sbjct: 305 ECCICLCSYE--DGAELHALPCNHHFHSSCIVKWLK--------MNATCPLCKYNI 442
>TC210241 weakly similar to UP|Q9SD55 (Q9SD55) Zinc-finger-like protein
(At3g47180), partial (21%)
Length = 928
Score = 32.0 bits (71), Expect = 0.40
Identities = 18/62 (29%), Positives = 26/62 (41%)
Frame = +3
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPK 112
+C CQ EEG+ + C H HT+C+ ++ CP C+ I PK
Sbjct: 531 RCVICQVEYEEGES--LVAIQCEHPYHTDCISKWLQ--------IKKVCPICNIEISAPK 680
Query: 113 SV 114
V
Sbjct: 681 MV 686
>BF009585 similar to GP|2970051|dbj ARG10 {Vigna radiata}, partial (57%)
Length = 469
Score = 31.6 bits (70), Expect = 0.53
Identities = 22/77 (28%), Positives = 35/77 (44%)
Frame = -3
Query: 86 HIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVS 145
H ++ HT +G CP C + PP++ + S +++ G+ K G HP +
Sbjct: 302 HWLTWTVHTVVSG--CPNC---LEPPQTSRISHP*QLMIWDATVIRCGVCKPTAGVHPTT 138
Query: 146 LSVTESRSPPPAFASEP 162
S RSP P A+ P
Sbjct: 137 ASPPTPRSPEPVSAASP 87
>TC219197
Length = 1042
Score = 31.2 bits (69), Expect = 0.69
Identities = 29/93 (31%), Positives = 38/93 (40%), Gaps = 19/93 (20%)
Frame = +1
Query: 40 SEWVI-DGEYDW--PPKCCQC-----------QAVLEEGDGSQTTR-----LGCLHVIHT 80
S+W+I G+ W P C QC Q+ L ++TT L LH T
Sbjct: 28 SQWLI*IGKQRWFTPTSCLQCPPNSLSRIITFQSQLCNSPFAKTTSPQRRLLSALHTTTT 207
Query: 81 NCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKS 113
+S S PP T+P G P+ S P KS
Sbjct: 208 FVSLSSEPSGPPRTSPIGPTSPS*SPPYTPSKS 306
>AW349923 weakly similar to PIR|T00747|T007 RING-H2 finger protein RHC1a
[imported] - Arabidopsis thaliana, partial (17%)
Length = 752
Score = 30.8 bits (68), Expect = 0.90
Identities = 17/63 (26%), Positives = 27/63 (41%), Gaps = 1/63 (1%)
Frame = -2
Query: 56 QCQAVLEEGD-GSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
QC EE + G + L C H+ H++C+V ++ +CP C + P S
Sbjct: 631 QCPVCQEEFEVGGEARELQCKHIYHSDCIVPWLR--------LHNSCPVCRHEVPVPSSS 476
Query: 115 KDS 117
S
Sbjct: 475 SSS 467
>TC216438 homologue to UP|Q8GUU2 (Q8GUU2) RES protein, partial (14%)
Length = 650
Score = 30.8 bits (68), Expect = 0.90
Identities = 17/56 (30%), Positives = 25/56 (44%)
Frame = +3
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
+CC C + E DG++ L C H H+ C+V +K CP C +I
Sbjct: 135 ECCICISSYE--DGAELHVLPCNHHFHSTCIVKWLK--------MNATCPLCKYNI 272
>TC216440 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (68%)
Length = 1655
Score = 30.8 bits (68), Expect = 0.90
Identities = 17/56 (30%), Positives = 25/56 (44%)
Frame = +1
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
+CC C + E DG++ L C H H+ C+V +K CP C +I
Sbjct: 1105 ECCICISSYE--DGAELHVLPCNHHFHSTCIVKWLK--------MNATCPLCKYNI 1242
>BU549221
Length = 326
Score = 29.6 bits (65), Expect = 2.0
Identities = 16/66 (24%), Positives = 30/66 (45%), Gaps = 1/66 (1%)
Frame = -2
Query: 56 QCQAVLEEGD-GSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
+C +E + GS+ ++ C H+ H++C+V + +CP C + P V
Sbjct: 220 KCSVXIERFEVGSEARKMPCDHIYHSDCIVPWLVHH--------NSCPVCRGKLPP*GHV 65
Query: 115 KDSGSR 120
GS+
Sbjct: 64 SSRGSQ 47
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,348,858
Number of Sequences: 63676
Number of extensions: 355260
Number of successful extensions: 2216
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 2177
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2205
length of query: 334
length of database: 12,639,632
effective HSP length: 98
effective length of query: 236
effective length of database: 6,399,384
effective search space: 1510254624
effective search space used: 1510254624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0218.25


