

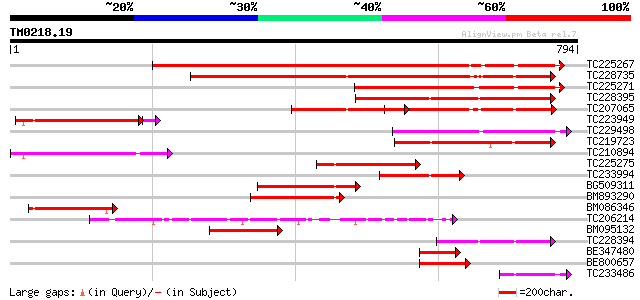
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.19
(794 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 603 e-173
TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (... 531 e-151
TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment... 258 9e-69
TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_3... 226 4e-59
TC207065 beta-glucosidase 220 2e-57
TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 204 1e-52
TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, parti... 198 7e-51
TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-li... 190 2e-48
TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%) 182 7e-46
TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylos... 167 1e-41
TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partia... 156 4e-38
BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 142 6e-34
BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 129 4e-30
BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabido... 127 2e-29
TC206214 beta-D-glucan exohydrolase [Glycine max] 119 5e-27
BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum v... 102 5e-22
TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 92 9e-19
BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus pe... 87 3e-17
BE800657 weakly similar to GP|4100433|gb|A beta-glucosidase {Gly... 76 7e-14
TC233486 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, parti... 62 1e-09
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 603 bits (1555), Expect = e-173
Identities = 306/580 (52%), Positives = 403/580 (68%), Gaps = 3/580 (0%)
Frame = +3
Query: 201 DRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKSCVVEGHVSSVMCS 260
+ LKV++CCKHYTAYD+DNW GVDRFHF+AKV+KQDLEDTY PFK+CV+EG V+SVMCS
Sbjct: 21 NHLKVAACCKHYTAYDLDNWNGVDRFHFNAKVSKQDLEDTYDVPFKACVLEGQVASVMCS 200
Query: 261 YNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATPEDAVALALKA 320
YN+VNG PTCADPDLL+ IRGQW L+GYIVSDCDSV V++++ HYT TPE+A A A+KA
Sbjct: 201 YNQVNGKPTCADPDLLRNTIRGQWRLNGYIVSDCDSVGVFFDNQHYTKTPEEAAAEAIKA 380
Query: 321 GLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLPFANLGPS 379
GL+++CG +L +T +A++ + + ++ AL V MRLG FD P + P+ NLGP
Sbjct: 381 GLDLDCGPFLAIHTDSAIRKGLISENDLNLALANLISVQMRLGMFDGEPSTQPYGNLGPR 560
Query: 380 DVCTKENQLLALEAAKQGIVLLENSG-VLPLSKTNIKNLAVIGPNANATTVMISNYAGIP 438
DVCT +Q LALEAA++ IVLL+N G LPLS + ++ + V+GPNA+AT MI NYAG+
Sbjct: 561 DVCTSAHQQLALEAARESIVLLQNKGNSLPLSPSRLRTIGVVGPNADATVTMIGNYAGVA 740
Query: 439 CRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAE 498
C YT+PLQG+ +Y+ + + GC V C LF AA A ADA+VLV+GLDQ++EAE
Sbjct: 741 CGYTTPLQGIARYVKT-AHQVGCKGVACRGNELFGAAETIARQADAIVLVMGLDQTVEAE 917
Query: 499 GLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQ 558
DRV L LPG Q++LV VA A KG VIL+IM+ GP+DISF K+ I ILWVGYPGQ
Sbjct: 918 TRDRVGLLLPGLQQELVTRVARAAKGPVILLIMSGGPVDISFAKNDPKISAILWVGYPGQ 1097
Query: 559 AGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNS 618
AGG AIA VIFG NPGGR P TWYPQ Y +VPMT+M+MR N + +PGRTYRFY G
Sbjct: 1098AGGTAIADVIFGTTNPGGRLPMTWYPQGYLAKVPMTNMDMRPNPTTGYPGRTYRFYKGPV 1277
Query: 619 LYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSDGQAIDISTISCLD-L 677
++ FGHGLSYS FS +A AP + + P S+ N+ S +A+ +S +C D L
Sbjct: 1278VFPFGHGLSYSRFSHSLALAPKQVSV-PIMSLQALTNSTLSS----KAVKVSHANCDDSL 1442
Query: 678 TFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEI 737
+ VKN G +G+H +L+F +PP + + IKQL+GF + HV+ G + V + +
Sbjct: 1443EMEFHVDVKNEGSMDGTHTLLIFSQPPHGK---WSQIKQLVGFHKTHVLAGSKQRVKVGV 1613
Query: 738 DICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQIDI 777
+C+ LS VD G R++ G+H L +G V+H I +
Sbjct: 1614HVCKHLSVVDQFGVRRIPTGEHELHIG-----DVKHSISV 1718
>TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (65%)
Length = 1599
Score = 531 bits (1367), Expect = e-151
Identities = 275/513 (53%), Positives = 365/513 (70%), Gaps = 2/513 (0%)
Frame = +1
Query: 254 VSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATPEDA 313
V+SVMCSYN+VNG PTCADPDLLKGV+RG+W L+GYIVSDCDSV+V Y+ HYT TPE+A
Sbjct: 1 VASVMCSYNKVNGKPTCADPDLLKGVVRGEWKLNGYIVSDCDSVEVLYKDQHYTKTPEEA 180
Query: 314 VALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLP 372
A+++ AGL++NCG +L +YT AVK +D ++++ A+ N+ LMRLGFFD +P+ P
Sbjct: 181 AAISILAGLDLNCGRFLGQYTEGAVKQGLIDEASINNAVTNNFATLMRLGFFDGDPRKQP 360
Query: 373 FANLGPSDVCTKENQLLALEAAKQGIVLLENSGV-LPLSKTNIKNLAVIGPNANATTVMI 431
+ NLGP DVCT+ENQ LA EAA+QGIVLL+NS LPL+ IK+LAVIGPNANAT VMI
Sbjct: 361 YGNLGPKDVCTQENQELAREAARQGIVLLKNSPASLPLNAKAIKSLAVIGPNANATRVMI 540
Query: 432 SNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGL 491
NY GIPC+Y SPLQGL + + +YA GC +V+C + L A K AA ADA V+VVG
Sbjct: 541 GNYEGIPCKYISPLQGLTAF-APTSYAAGCLDVRCPNPVL-DDAKKIAASADATVMVVGA 714
Query: 492 DQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGIL 551
+IEAE LDRVN+ LPG Q+ LV +VA+A+KG VILVIM+ G +D+SF K+ I IL
Sbjct: 715 SLAIEAESLDRVNILLPGQQQLLVSEVANASKGPVILVIMSGGGMDVSFAKNNNKITSIL 894
Query: 552 WVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTY 611
WVGYPG+AGG AIA VIFG +NP GR P TWYPQSY D+VPMT+MNMR + + +PGRTY
Sbjct: 895 WVGYPGEAGGAAIADVIFGFHNPSGRLPMTWYPQSYVDKVPMTNMNMRPDPATGYPGRTY 1074
Query: 612 RFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSDGQAIDIST 671
RFY G +++ FG GLSYS+ + AP + ++ +++ H + S S+ ++ID+
Sbjct: 1075RFYKGETVFAFGDGLSYSSIVHKLVKAPQLVSVQ----LAEDH--VCRS-SECKSIDVVG 1233
Query: 672 ISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTE 731
C +L F + + +KN G + +H V +F PP V AP K L+GFE+ H++
Sbjct: 1234EHCQNLVFDIHLRIKNKGKMSSAHTVFLFSTPPA---VHNAPQKHLLGFEKVHLIGKSEA 1404
Query: 732 FVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVG 764
V+ ++D+C+ LS VD G RK+ +G+H L VG
Sbjct: 1405LVSFKVDVCKDLSIVDELGNRKVALGQHLLHVG 1503
>TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment), partial
(55%)
Length = 952
Score = 258 bits (658), Expect = 9e-69
Identities = 138/295 (46%), Positives = 187/295 (62%), Gaps = 1/295 (0%)
Frame = +3
Query: 484 AVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKS 543
A VLV+GLDQ+IEAE DRV L LPG Q++LV VA A KG VILVIM+ GP+D+SF K+
Sbjct: 3 ATVLVMGLDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKN 182
Query: 544 IRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSS 603
I ILWVGYPGQAGG AIA VIFG NPGGR P TWYPQ Y +VPMT+M+MR N +
Sbjct: 183 NPKISAILWVGYPGQAGGTAIADVIFGATNPGGRLPMTWYPQGYLAKVPMTNMDMRPNPA 362
Query: 604 RNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSD 663
+PGRTYRFY G ++ FGHGLSYS FS +A AP + ++ + + + S
Sbjct: 363 TGYPGRTYRFYKGPVVFPFGHGLSYSRFSQSLALAPKQVSVQILSLQA-----LTNSTLS 527
Query: 664 GQAIDISTISCLD-LTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFER 722
+A+ +S +C D L + VKN G +G+H +L+F +PP + + IKQL+ F +
Sbjct: 528 SKAVKVSHANCDDSLETEFHVDVKNEGSMDGTHTLLIFSKPPPGK---WSQIKQLVTFHK 698
Query: 723 AHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQIDI 777
HV G + + + + C+ LS VD G R++ G+H L +G ++H I++
Sbjct: 699 THVPAGSKQRLKVNVHSCKHLSVVDQFGVRRIPTGEHELHIG-----DLKHSINV 848
>TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial
(22%)
Length = 1011
Score = 226 bits (575), Expect = 4e-59
Identities = 121/282 (42%), Positives = 175/282 (61%), Gaps = 2/282 (0%)
Frame = +3
Query: 485 VVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSI 544
VVLV+GLDQS E E DR L LPG QE+L+K VA A+K V+LV++ GP+DI+ K
Sbjct: 18 VVLVMGLDQSQERESHDREYLGLPGKQEELIKSVARASKRPVVLVLLCGGPVDITSAKFD 197
Query: 545 RNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSR 604
+GGILW GYPG+ GG A+AQV+FG++NPGG+ P TWYP+ + +VPMTDM MRA+ +
Sbjct: 198 DKVGGILWAGYPGELGGVALAQVVFGDHNPGGKLPITWYPKDFI-KVPMTDMRMRADPAS 374
Query: 605 NFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIAS-APSTIMIEPNTSMSQPHNNIFESLSD 663
+PGRTYRFY G +YEFG+GLSY+ +S + S + ST+ I +++ N+ E++
Sbjct: 375 GYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLLSLSHSTLHINQSSTHLMTQNS--ETIRY 548
Query: 664 GQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVF-WEPPTSELVTGAPIKQLIGFER 722
+++ +C + + +GV N G G H VL+F + + G P+KQL+GF+
Sbjct: 549 KLVSELAEETCQTMLLSIALGVTNRGNLAGKHPVLLFVRQGKVRNINNGNPVKQLVGFQS 728
Query: 723 AHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVG 764
V G T V E+ C+ LS + G + G + +VG
Sbjct: 729 VKVNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLFIVG 854
>TC207065 beta-glucosidase
Length = 1328
Score = 220 bits (561), Expect = 2e-57
Identities = 114/240 (47%), Positives = 157/240 (64%)
Frame = +2
Query: 526 VILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQ 585
VILVIM+ G +D+SF KS I ILWVGYPG+AGG AIA VIFG YNP GR P TWYPQ
Sbjct: 488 VILVIMSGGGMDVSFAKSNDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPQ 667
Query: 586 SYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIE 645
+Y ++VPMT+MNMRA+ + +PGRTYRFY G +++ FG G+S+S+ I AP + +
Sbjct: 668 AYVNKVPMTNMNMRADPATGYPGRTYRFYKGETVFSFGDGISFSSIEHKIVKAPQLVSV- 844
Query: 646 PNTSMSQPHNNIFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPT 705
+++ H S+ ++DI+ C +L F + +GVKNTG + SHVVL+F+ PP
Sbjct: 845 ---PLAEDHEC---RSSECMSLDIADEHCQNLAFDIHLGVKNTGKMSTSHVVLLFFTPPD 1006
Query: 706 SELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGS 765
V AP K L+GFE+ H+ V ++D+C+ LS VD G RK+ +G+H L VG+
Sbjct: 1007---VHNAPQKHLLGFEKVHLPGKSEAQVRFKVDVCKDLSVVDELGNRKVPLGQHLLHVGN 1177
Score = 180 bits (456), Expect = 2e-45
Identities = 97/166 (58%), Positives = 121/166 (72%), Gaps = 1/166 (0%)
Frame = +1
Query: 395 KQGIVLLENS-GVLPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYIS 453
+QGIVLL+NS G LPL+ IK+LAVIGPNANAT VMI NY GIPC Y SPLQ L +
Sbjct: 4 RQGIVLLKNSPGSLPLNAKTIKSLAVIGPNANATRVMIGNYEGIPCNYISPLQTLTALVP 183
Query: 454 SVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEK 513
+ +YA GC NV+C + L AT+ AA ADA V++VG +IEAE LDR+N+ LPG Q+
Sbjct: 184 T-SYAAGCPNVQCANAEL-DDATQIAASADATVIIVGASLAIEAESLDRINILLPGQQQL 357
Query: 514 LVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQA 559
LV +VA+A+KG VILVIM+ G +D+SF KS I ILW+GYPG +
Sbjct: 358 LVSEVANASKGPVILVIMSGGGMDVSFAKSNDKITSILWIGYPGDS 495
>TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (20%)
Length = 638
Score = 204 bits (520), Expect(2) = 1e-52
Identities = 100/183 (54%), Positives = 133/183 (72%), Gaps = 4/183 (2%)
Frame = +3
Query: 9 SLFLSLLVLL---PILTSSQKHACNKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKA 65
++F+S L+L ++ ++C+ +S + +PFC+T L RA+DLVSRLTL EK
Sbjct: 18 AIFISFLLLTLHHHAESTQPPYSCDSSS-NSPYYPFCNTRLPITKRAQDLVSRLTLDEKL 194
Query: 66 QQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNAT 125
QLVN + I RLG+PSY+WWSEALHGV++ G G RFN T+ ATSFP VIL+AASF+
Sbjct: 195 AQLVNTAPAIPRLGIPSYQWWSEALHGVADAGFGIRFNGTIKSATSFPQVILTAASFDPN 374
Query: 126 LWYNMGQVVSTEARAMYNVDLA-GLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVK 184
LWY + + + EARA+YN A G+TFW+PN+NVFRDPRWGRGQET GEDPL+ ++Y V
Sbjct: 375 LWYQISKTIGKEARAVYNAGQATGMTFWAPNINVFRDPRWGRGQETAGEDPLMNAKYGVA 554
Query: 185 YVR 187
YV+
Sbjct: 555 YVK 563
Score = 21.2 bits (43), Expect(2) = 1e-52
Identities = 10/25 (40%), Positives = 12/25 (48%)
Frame = +1
Query: 187 RGLQDVEDEESIKADRLKVSSCCKH 211
R LQ E +RL +CCKH
Sbjct: 562 RXLQGDSFEGGKLGERL*AXACCKH 636
>TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial (24%)
Length = 792
Score = 198 bits (504), Expect = 7e-51
Identities = 105/250 (42%), Positives = 148/250 (59%)
Frame = +1
Query: 537 DISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDM 596
DI+F K+ I GILW GYPGQAGG AIA ++FG NPGG+ P TWYP+ Y ++PMT+M
Sbjct: 1 DITFAKNNPRIVGILWAGYPGQAGGAAIADILFGTANPGGKLPVTWYPEEYLTKLPMTNM 180
Query: 597 NMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNN 656
MRA S +PGRTYRFYNG +Y FGHGL+Y+ F +ASAP+ + + N N
Sbjct: 181 AMRATKSAGYPGRTYRFYNGPVVYPFGHGLTYTHFVHTLASAPTVVSVPLNGHRRANVTN 360
Query: 657 IFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQ 716
I +AI ++ C L+ L + +KN G +G+H +LVF PP A KQ
Sbjct: 361 I-----SNRAIRVTHARCDKLSISLEVDIKNVGSRDGTHTLLVFSAPPAG-FGHWALEKQ 522
Query: 717 LIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQID 776
L+ FE+ HV + V + I +C+LLS VD G R++ +G+H+ +G V+H +
Sbjct: 523 LVAFEKIHVPAKGLQRVGVNIHVCKLLSVVDKSGIRRIPLGEHSFNIG-----DVKHSVS 687
Query: 777 ISLSGNGMEK 786
+ + G+ K
Sbjct: 688 LQAAALGIIK 717
>TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-like protein
(AT5g10560/F12B17_90), partial (19%)
Length = 1013
Score = 190 bits (482), Expect = 2e-48
Identities = 97/229 (42%), Positives = 140/229 (60%), Gaps = 4/229 (1%)
Frame = +1
Query: 540 FTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMR 599
F + I I+W+GYPG+AGG A+A++IFGE+NP GR P TWYP+++ + VPM +M+MR
Sbjct: 1 FAEKNPQIASIIWLGYPGEAGGKALAEIIFGEFNPAGRLPMTWYPEAFTN-VPMNEMSMR 177
Query: 600 ANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFE 659
A+ SR +PGRTYRFY G +Y FGHGLS+S FS SAPS I + +++
Sbjct: 178 ADPSRGYPGRTYRFYTGGRVYGFGHGLSFSDFSYNFLSAPSKISLSRTIKDGSRKRLLYQ 357
Query: 660 SLSDGQAIDISTI----SCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIK 715
++ +D + +C L+F + I V N G +GSHVV++F + P ++V G+P
Sbjct: 358 VENEVYGVDYVPVNQLQNCNKLSFSVHISVMNLGGLDGSHVVMLFSKGP--KVVDGSPET 531
Query: 716 QLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVG 764
QL+GF R H + ++ + C+ LS D GKR L +G HTL VG
Sbjct: 532 QLVGFSRLHTISSKPTETSILVHPCEHLSFADKQGKRILPLGPHTLSVG 678
>TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%)
Length = 738
Score = 182 bits (461), Expect = 7e-46
Identities = 105/230 (45%), Positives = 133/230 (57%), Gaps = 4/230 (1%)
Frame = +3
Query: 2 KSNLLFLSLFLSLLVLL---PILTSSQ-KHACNKASPKTSNFPFCDTTLSYEARAKDLVS 57
K L L+L L L V L P +T + AC+ + T F FC+T + R +DL++
Sbjct: 51 KKGHLLLALGLVLCVTLTFFPRVTEGRVPFACDPRNGLTRGFKFCNTHVPIHVRVQDLIA 230
Query: 58 RLTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVIL 117
RLTL EK + +VN + + RLG+ YEWWSEALHGVSN+GPGT+F PGAT FP VI
Sbjct: 231 RLTLPEKIRLVVNNAIAVPRLGIQGYEWWSEALHGVSNVGPGTKFGGAFPGATMFPQVIS 410
Query: 118 SAASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLV 177
+AASFN +LW +G+VVS EARAMYN AGLT+WSPNVN+FRDP G P +
Sbjct: 411 TAASFNQSLWQEIGRVVSDEARAMYNGGQAGLTYWSPNVNIFRDPPVGPWPRDSRRGPDI 590
Query: 178 VSRYAVKYVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFH 227
K R L ++ + CKHY A + GVDRFH
Sbjct: 591 ----GRKIRRQLCQRTPRRWCPEIASRLLAWCKHYMAMILIIGNGVDRFH 728
>TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (32%)
Length = 933
Score = 167 bits (424), Expect = 1e-41
Identities = 86/146 (58%), Positives = 101/146 (68%)
Frame = +3
Query: 430 MISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVV 489
MI NYAG+ C YT+PLQG+ +Y+ + + GC V C LF AA A DA VLV+
Sbjct: 12 MIGNYAGVACGYTTPLQGIARYVKTA-HQVGCRGVACRGNELFGAAEIIARQVDATVLVM 188
Query: 490 GLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGG 549
GLDQ+IEAE DRV L LPG Q++LV VA A KG VILVIM+ GP+D+SF K+ I
Sbjct: 189 GLDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKNNPKISA 368
Query: 550 ILWVGYPGQAGGDAIAQVIFGEYNPG 575
ILWVGYPGQAGG AIA VIFG NPG
Sbjct: 369 ILWVGYPGQAGGTAIADVIFGATNPG 446
>TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (15%)
Length = 462
Score = 156 bits (394), Expect = 4e-38
Identities = 72/120 (60%), Positives = 92/120 (76%)
Frame = +2
Query: 518 VADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGR 577
VA+A+K VILV+++ GP+DI+ K IGGILW GYPG+ GG A+AQ+IFG++NPGGR
Sbjct: 2 VAEASKKPVILVLLSGGPLDITSAKYNHKIGGILWAGYPGELGGIALAQIIFGDHNPGGR 181
Query: 578 SPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIAS 637
P TWYP+ Y +VPMTDM MRA+ S +PGRTYRFY G +YEFG+GLSYS +S + S
Sbjct: 182 LPTTWYPKDYI-KVPMTDMRMRADPSTGYPGRTYRFYKGPKVYEFGYGLSYSKYSYELVS 358
>BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (31%)
Length = 443
Score = 142 bits (358), Expect = 6e-34
Identities = 80/146 (54%), Positives = 99/146 (67%), Gaps = 2/146 (1%)
Frame = +1
Query: 348 VDQALVYNYIVLMRLGFFDN-PKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSG- 405
V+ ALV V MRLG FD P + P+ +LGP DVC +Q LALEAA+QGIVLL+N+G
Sbjct: 7 VNGALVNTLTVQMRLGMFDGEPTAHPYGHLGPKDVCKPAHQELALEAARQGIVLLKNTGP 186
Query: 406 VLPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVK 465
VLPLS + +AVIGPN+ AT MI NYAG+ C YT+PLQG+ +Y +V + GC NV
Sbjct: 187 VLPLSSQLHRTVAVIGPNSKATITMIGNYAGVACGYTNPLQGIGRYARTV-HQLGCQNVA 363
Query: 466 CGDQSLFAAATKAAAPADAVVLVVGL 491
C + LF A AA ADA VLV+GL
Sbjct: 364 CKNDKLFGPAINAARQADATVLVMGL 441
>BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (27%)
Length = 403
Score = 129 bits (325), Expect = 4e-30
Identities = 72/133 (54%), Positives = 88/133 (66%), Gaps = 2/133 (1%)
Frame = +1
Query: 338 VKLKKVDVSTVDQALVYNYIVLMRLGFFDN-PKSLPFANLGPSDVCTKENQLLALEAAKQ 396
VK + + V+ AL+ V MRLG +D P S P+ NLGP DVCT+ +Q LALEAA+Q
Sbjct: 1 VKKGLISEADVNGALLNTLTVQMRLGMYDGEPSSHPYNNLGPRDVCTQSHQELALEAARQ 180
Query: 397 GIVLLENSGV-LPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSV 455
GIVLL+N G LPLS + +AVIGPN+N T MI NYAGI C YTSPLQG+ Y ++
Sbjct: 181 GIVLLKNKGPSLPLSTRRGRTVAVIGPNSNVTFTMIGNYAGIACGYTSPLQGIGTYTKTI 360
Query: 456 TYAPGCGNVKCGD 468
Y GC NV C D
Sbjct: 361 -YEHGCANVACTD 396
>BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabidopsis
thaliana}, partial (11%)
Length = 427
Score = 127 bits (319), Expect = 2e-29
Identities = 65/129 (50%), Positives = 87/129 (67%), Gaps = 5/129 (3%)
Frame = -3
Query: 27 HACNKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKAQQLVNPSSGIARLGVPSYEWW 86
++C+ +S + +PFC+T L RA+DLVSRLTL EK QLVN + I RLG+PSY+WW
Sbjct: 407 YSCDSSS-NSPYYPFCNTRLPITKRAQDLVSRLTLDEKLAQLVNTAPAIPRLGIPSYQWW 231
Query: 87 SEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVV-----STEARAM 141
SEALHGV++ G G RFN T+ ATSFP VIL+AASF+ LWY + +V +A+
Sbjct: 230 SEALHGVADAGFGIRFNGTIKSATSFPQVILTAASFDPNLWYQISKVSYIITNIQHLKAL 51
Query: 142 YNVDLAGLT 150
Y + L +T
Sbjct: 50 YTLQLLFIT 24
>TC206214 beta-D-glucan exohydrolase [Glycine max]
Length = 1647
Score = 119 bits (298), Expect = 5e-27
Identities = 140/552 (25%), Positives = 241/552 (43%), Gaps = 36/552 (6%)
Frame = +1
Query: 112 FPAVILSAASFNATLWYNMGQVVSTEARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQET 170
FP + + + L +G+ + E RA G+ + ++P + V RDPRWGR E+
Sbjct: 4 FPHNVGLGVTRDPVLIKKIGEATALEVRA------TGIPYVFAPCIAVCRDPRWGRCYES 165
Query: 171 PGEDPLVVSRYAVKYVRGLQDVEDEESIK-----ADRLKVSSCCKHYTAYDVDNWKGVDR 225
EDP +V + + + GLQ SIK A + KV++C KHY D KG++
Sbjct: 166 YSEDPKIV-KTMTEIIPGLQGDIPGNSIKGVPFVAGKNKVAACAKHYLG-DGGTNKGIN- 336
Query: 226 FHFDAKVTKQDLEDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWG 285
+ ++ L + P + +++G VS+VM SY+ NG+ A+ L+ G ++ +
Sbjct: 337 -ENNTLISYNGLLSIHMPAYYDSIIKG-VSTVMISYSSWNGMKMHANKKLITGYLKNKLH 510
Query: 286 LDGYIVSDCDSVQVYYESIHYTATPEDAVALALKAGLNM-----NCGDYLRKYTANAVKL 340
G+++SD + H A +V + AG++M N +++ + T VK
Sbjct: 511 FKGFVISDWQGIDRITSPPH--ANYSYSVQAGVSAGIDMIMVPFNYTEFIDELT-RQVKN 681
Query: 341 KKVDVSTVDQALVYNYIVLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVL 400
+ +S +D A+ V +G F+NP + P + + +KE++ +A EA ++ +VL
Sbjct: 682 NIIPISRIDDAVARILRVKFVMGLFENPYADPSL---ANQLGSKEHREIAREAVRKSLVL 852
Query: 401 LEN-----SGVLPLSKTNIKNLAVIGPNANATTVMISNYAGIPC-RYTSPLQGLQKYISS 454
L+N +LPL K + K + V G +A N G C +T QGL
Sbjct: 853 LKNGKSYKKPLLPLPKKSTK-ILVAGSHA--------NNLGYQCGGWTITWQGLG----- 990
Query: 455 VTYAPGCGNVKCGDQSLFAAATKAAAPA-------------------DAVVLVVGLDQSI 495
GN ++ A + PA D ++VVG
Sbjct: 991 -------GNDLTSGTTILDAVKQTVDPATEVVFNENPDKNFVKSYKFDYAIVVVGEHTYA 1149
Query: 496 EAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGY 555
E G D +NLT+ + +V A + ++V++ P+ I + I ++
Sbjct: 1150ETFG-DSLNLTMADPGPSTITNVCGAI--RCVVVLVTGRPVVIK--PYLPKIDALVAAWL 1314
Query: 556 PGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYN 615
PG G +A V++G+Y G+ TW+ DQ+PM N + Y
Sbjct: 1315PG-TEGQGVADVLYGDYEFTGKLARTWF--KTVDQLPM-----------NVGDKHY---- 1440
Query: 616 GNSLYEFGHGLS 627
+ L+ FG+GL+
Sbjct: 1441-DPLFPFGYGLT 1473
>BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum vulgare},
partial (2%)
Length = 428
Score = 102 bits (255), Expect = 5e-22
Identities = 47/103 (45%), Positives = 68/103 (65%), Gaps = 1/103 (0%)
Frame = +1
Query: 281 RGQWGLDGYIVSDCDSVQVYYESIHYTATPEDAVALALKAGLNMNCGDYLRKYTANAVKL 340
R QW DGYI SDC +V + +E Y T EDA+A +AG+++ CGDY+ K+ +AV
Sbjct: 13 RQQWKFDGYITSDCGAVSIIHEKQGYAKTAEDAIADVFRAGMDVECGDYITKHAKSAVFQ 192
Query: 341 KKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLPFANLGPSDVC 382
KK+ +S +D+AL + + +RLG FD NP LPF +GP++VC
Sbjct: 193 KKLPISQIDRALQNLFSIRIRLGLFDGNPTKLPFGTIGPNEVC 321
>TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (14%)
Length = 659
Score = 92.0 bits (227), Expect = 9e-19
Identities = 56/168 (33%), Positives = 90/168 (53%), Gaps = 1/168 (0%)
Frame = +2
Query: 598 MRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIAS-APSTIMIEPNTSMSQPHNN 656
MRA+ + +PGRTYRFY G +YEFG+GLSY+ +S + S + +T+ I +++ N+
Sbjct: 2 MRADPASGYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLLSLSHNTLHINQSSTHLTTQNS 181
Query: 657 IFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQ 716
E++ +++ +C + + +GV N G G H VL+F G P+KQ
Sbjct: 182 --ETIRYKLVSELAEETCQTMLLSIALGVTNHGNMAGKHPVLLFVRQGKVR-NNGNPVKQ 352
Query: 717 LIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVG 764
L+GF+ + G T V E+ C+ LS + G + G + LLVG
Sbjct: 353 LVGFQSVKLNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLLLVG 496
>BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus persica},
partial (17%)
Length = 473
Score = 87.0 bits (214), Expect = 3e-17
Identities = 36/58 (62%), Positives = 45/58 (77%)
Frame = +3
Query: 574 PGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTF 631
PGG+ P TWYPQ Y +PMT+M MRA+ S+ +PGRTYRFYNG +Y FG+GLSY+ F
Sbjct: 282 PGGKLPMTWYPQGYIKNLPMTNMAMRASRSKGYPGRTYRFYNGPVVYPFGYGLSYTHF 455
Score = 34.3 bits (77), Expect = 0.22
Identities = 14/20 (70%), Positives = 16/20 (80%)
Frame = +2
Query: 556 PGQAGGDAIAQVIFGEYNPG 575
PGQAGG AIA ++FG NPG
Sbjct: 2 PGQAGGAAIADILFGTSNPG 61
>BE800657 weakly similar to GP|4100433|gb|A beta-glucosidase {Glycine max},
partial (19%)
Length = 220
Score = 75.9 bits (185), Expect = 7e-14
Identities = 33/72 (45%), Positives = 46/72 (63%)
Frame = +2
Query: 574 PGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFST 633
PGGR P TWYPQ Y +VP+T+++M N S +P +TYRFY ++ F H LSYS FS
Sbjct: 2 PGGRLPMTWYPQRYLAKVPITNIDMHPNPSTRYP*KTYRFYK*PVIFPFKHKLSYSRFSQ 181
Query: 634 YIASAPSTIMIE 645
++ P + I+
Sbjct: 182 ILSLTPKQVSIQ 217
>TC233486 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial (7%)
Length = 677
Score = 62.0 bits (149), Expect = 1e-09
Identities = 35/101 (34%), Positives = 55/101 (53%)
Frame = +2
Query: 686 KNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSN 745
KN G +G+H +LVF PP AP KQL+ F++ H+ + V + I +C+LLS
Sbjct: 2 KNVGSKDGTHTLLVFSAPPAGN-GHWAPHKQLVAFQKLHIPSKAQQRVNVNIHVCKLLSV 178
Query: 746 VDSDGKRKLIIGKHTLLVGSSSETQVRHQIDISLSGNGMEK 786
VD G R++ +G H+L +G V+H + + G+ K
Sbjct: 179 VDRSGTRRVPMGLHSLHIG-----DVKHYVSLQAETLGIIK 286
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,315,656
Number of Sequences: 63676
Number of extensions: 453578
Number of successful extensions: 2301
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 2245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2260
length of query: 794
length of database: 12,639,632
effective HSP length: 105
effective length of query: 689
effective length of database: 5,953,652
effective search space: 4102066228
effective search space used: 4102066228
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0218.19


