

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.16
(560 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
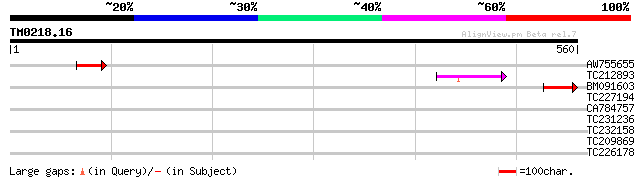
Score E
Sequences producing significant alignments: (bits) Value
AW755655 57 2e-08
TC212893 similar to UP|Q84VX1 (Q84VX1) At4g38650, partial (17%) 52 7e-07
BM091603 47 2e-05
TC227194 similar to UP|Q8T5S8 (Q8T5S8) Prolyl 4-hydroxylase alph... 40 0.003
CA784757 31 1.3
TC231236 31 1.3
TC232158 30 2.8
TC209869 similar to UP|Q9LG03 (Q9LG03) F20N2.6, partial (33%) 30 3.7
TC226178 UP|Q8BU53 (Q8BU53) Mus musculus 2 days pregnant adult f... 28 8.3
>AW755655
Length = 145
Score = 57.4 bits (137), Expect = 2e-08
Identities = 26/29 (89%), Positives = 27/29 (92%)
Frame = +3
Query: 67 ARNRTKPLDSFSQKVQLKKGMIYTFSAWL 95
A NRT PLDSFSQKVQLKKGM+YTFSAWL
Sbjct: 51 AHNRTHPLDSFSQKVQLKKGMLYTFSAWL 137
>TC212893 similar to UP|Q84VX1 (Q84VX1) At4g38650, partial (17%)
Length = 609
Score = 52.0 bits (123), Expect = 7e-07
Identities = 29/72 (40%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Frame = +3
Query: 422 LDILGATGYPIWLTETSIDP---QPNQEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQK 478
LD L G PIWLTE I + Q Y E +LREG+SHP+V GI+++ G +
Sbjct: 9 LDKLATLGLPIWLTEVDISKTLDRDAQANYLEEVLREGFSHPSVNGIMLWTAFHPNGCYQ 188
Query: 479 APLADENFTNTP 490
L D F P
Sbjct: 189 MCLTDNYFQEPP 224
Score = 41.2 bits (95), Expect = 0.001
Identities = 27/85 (31%), Positives = 35/85 (40%)
Frame = +1
Query: 472 VQAGFQKAPLADENFTNTPMGDVVDKLIREWGTGPHETMADSRGIVDISLHHGDYDVIVS 531
+Q G K F N P GDVVDKL+ EW E + D G G+Y + V
Sbjct: 169 IQMGATKCA*QTIIFRNLPAGDVVDKLVEEWQISRVEGVTDVHGSYSFYGFLGEYRISVK 348
Query: 532 HPQTHFSKTLNLSVRKEFSQQTIHV 556
+ T +LS E TI +
Sbjct: 349 YGNKTTKSTFSLSRGDETRHFTITI 423
>BM091603
Length = 198
Score = 47.4 bits (111), Expect = 2e-05
Identities = 22/33 (66%), Positives = 26/33 (78%)
Frame = +2
Query: 528 VIVSHPQTHFSKTLNLSVRKEFSQQTIHVKMHA 560
V V+HP H TLNLSV+K+FS +TIHVKMHA
Sbjct: 2 VTVTHPLIHSPITLNLSVKKDFSLETIHVKMHA 100
>TC227194 similar to UP|Q8T5S8 (Q8T5S8) Prolyl 4-hydroxylase alpha-related
protein PH4[alpha]PV (CG31015-PA), partial (4%)
Length = 965
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/80 (28%), Positives = 36/80 (44%), Gaps = 2/80 (2%)
Frame = +1
Query: 320 DLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANP 379
D I D+ HY+F GE ++Y +P+ FL++FNT + G +A
Sbjct: 130 DKIVRDIEKRIAHYSFIPVEHGEGLQVLHYEVGQKYEPHYDYFLDDFNT-KNGGQRIATV 306
Query: 380 ANYIRKIREIQQ--FPGTKG 397
Y+ + E + FP KG
Sbjct: 307 LMYLTDVEEGGETVFPAAKG 366
>CA784757
Length = 409
Score = 31.2 bits (69), Expect = 1.3
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = +1
Query: 408 FASGNPNLAYMRSGLDILGATGYPIWLTETSIDP 441
F+ P LA+ R+GL + TG+P WL S+ P
Sbjct: 292 FSGPRPQLAWFRNGLRL---TGFPFWLRSFSLGP 384
>TC231236
Length = 1393
Score = 31.2 bits (69), Expect = 1.3
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 7/78 (8%)
Frame = +2
Query: 23 PQREQYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKPLD-----SF 77
P+ + G I+ P + + WT+ G K +G+ +V + T + S
Sbjct: 146 PKPSELKGSIVTTP---NGIPHWTISGMVEYIKSGQKQGDMVLVVPHGTYAVRLGNEASI 316
Query: 78 SQKVQLKKGMIY--TFSA 93
QK+Q+ KGM Y TFSA
Sbjct: 317 KQKIQVVKGMFYSLTFSA 370
>TC232158
Length = 904
Score = 30.0 bits (66), Expect = 2.8
Identities = 20/91 (21%), Positives = 39/91 (41%)
Frame = +2
Query: 298 SPQDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDP 357
S Q + RK + + R + +++ ++ F GE ++Y + DP
Sbjct: 329 SGQSVVHDVRKSTGAFLDRGQDEIVR-NIEKRIADVTFIPIENGEPIYVIHYEVGQYYDP 505
Query: 358 NARMFLNEFNTIEYSGDVVANPANYIRKIRE 388
+ F+++FN IE G +A Y+ + E
Sbjct: 506 HYDYFIDDFN-IENGGQRIATMLMYLSNVEE 595
>TC209869 similar to UP|Q9LG03 (Q9LG03) F20N2.6, partial (33%)
Length = 1085
Score = 29.6 bits (65), Expect = 3.7
Identities = 15/63 (23%), Positives = 25/63 (38%)
Frame = +3
Query: 231 KWFMSRFKHTTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQ 290
KW SRF H +++ W + V +++ + NG G + + DR Q
Sbjct: 57 KWIFSRFSHFQYSSSCSWQGGQTVCSSQSFKSHER-------NGYRTNGASFHYIDRWTQ 215
Query: 291 PEW 293
W
Sbjct: 216 QSW 224
>TC226178 UP|Q8BU53 (Q8BU53) Mus musculus 2 days pregnant adult female oviduct
cDNA, RIKEN full-length enriched library,
clone:E230038K10 product:procollagen-proline,
2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase),
alpha II polypeptide, full insert sequence, partial (8%)
Length = 1637
Score = 28.5 bits (62), Expect = 8.3
Identities = 14/52 (26%), Positives = 25/52 (47%)
Frame = +1
Query: 302 LREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAY 353
+R AA + + + DL DV+N NLH N+ +F + +T++
Sbjct: 1054 MRTAALPTFRKLYGKIEVDLEKGDVINVNLHNNYNTYSFNGKKKLVLSTTSF 1209
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,937,384
Number of Sequences: 63676
Number of extensions: 352788
Number of successful extensions: 1534
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1528
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1533
length of query: 560
length of database: 12,639,632
effective HSP length: 102
effective length of query: 458
effective length of database: 6,144,680
effective search space: 2814263440
effective search space used: 2814263440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0218.16


