

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.11
(360 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
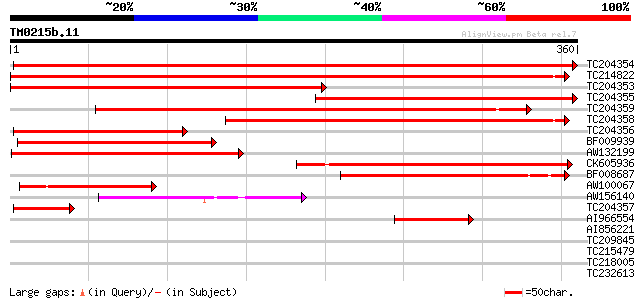
Score E
Sequences producing significant alignments: (bits) Value
TC204354 similar to UP|Q852S8 (Q852S8) S-adenosylmethionine deca... 609 e-175
TC214822 UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase, ... 476 e-135
TC204353 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine de... 283 1e-76
TC204355 similar to UP|Q852S8 (Q852S8) S-adenosylmethionine deca... 282 1e-76
TC204359 similar to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine deca... 275 2e-74
TC204358 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine de... 259 2e-69
TC204356 similar to UP|Q6KC47 (Q6KC47) S-adenosylmethionine deca... 194 5e-50
BF009939 weakly similar to GP|18152533|emb S-adenosyl-L-methioni... 160 6e-40
AW132199 similar to GP|19526455|gb S-adenosylmethionine decarbox... 157 5e-39
CK605936 145 4e-35
BF008687 132 2e-31
AW100067 similar to GP|21239731|gb| S-adenosylmethionine decarbo... 115 4e-26
AW156140 weakly similar to GP|19526455|gb| S-adenosylmethionine ... 83 2e-16
TC204357 similar to UP|Q6KC47 (Q6KC47) S-adenosylmethionine deca... 69 4e-12
AI966554 similar to GP|19526455|gb| S-adenosylmethionine decarbo... 58 8e-09
AI856221 37 0.018
TC209845 similar to PIR|E87912|E87912 protein B0205.8 [imported]... 30 2.2
TC215479 29 2.9
TC218005 similar to UP|Q8RVB2 (Q8RVB2) SPY protein, partial (19%) 29 3.8
TC232613 similar to UP|Q8LEH1 (Q8LEH1) Ripening-related protein-... 28 8.4
>TC204354 similar to UP|Q852S8 (Q852S8) S-adenosylmethionine decarboxylase,
partial (83%)
Length = 1790
Score = 609 bits (1570), Expect = e-175
Identities = 296/359 (82%), Positives = 331/359 (91%), Gaps = 1/359 (0%)
Frame = +3
Query: 3 LTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDY 62
+ SAIGFEGYEKRLEISF+E GVFADPEG+GLR LS+DQLDEIL PA+C+IV SLSNDY
Sbjct: 462 MAGSAIGFEGYEKRLEISFFEHGVFADPEGIGLRALSKDQLDEILKPAECSIVASLSNDY 641
Query: 63 VDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPE 122
VDSYVLSESSLF+YPYK+IIKTCGTTKLLLSIPVILKLAD+L+IAVKSVRYTRGSFIFP
Sbjct: 642 VDSYVLSESSLFIYPYKIIIKTCGTTKLLLSIPVILKLADALDIAVKSVRYTRGSFIFPG 821
Query: 123 AQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGL 182
AQ FPHR+FSEEV+VL++YF NLGSG +AYVMGDP KSQIWHIY+ASA+ KGSSEAVYGL
Sbjct: 822 AQSFPHRSFSEEVSVLDSYFSNLGSGSKAYVMGDPSKSQIWHIYSASAQTKGSSEAVYGL 1001
Query: 183 EMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSA 242
EMCMTGL+KESASVFFK+NTSSAALMT NSGI KILPQSDI DFEFDPCGYSMNGIEG A
Sbjct: 1002EMCMTGLEKESASVFFKENTSSAALMTENSGIRKILPQSDISDFEFDPCGYSMNGIEGGA 1181
Query: 243 ISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDK 302
ISTIHVTPEDGFSYASFEAVGYD+++++L ELVERVLACF PAEFSVALH+DMHGEKL+K
Sbjct: 1182ISTIHVTPEDGFSYASFEAVGYDFDEMALGELVERVLACFRPAEFSVALHIDMHGEKLNK 1361
Query: 303 FPLDIEGYYCDKRGTEELGAGGAVMFHSFVR-VDGCASPKSILKCCWSEDENDEEVKGI 360
FPLDI+GYYC +R EELG GGAV++ SFV+ DG SP+SILKCCWSEDEN++EV+ I
Sbjct: 1362FPLDIKGYYCGERSYEELGVGGAVVYQSFVQGCDGSTSPRSILKCCWSEDENEDEVREI 1538
>TC214822 UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase, complete
Length = 1826
Score = 476 bits (1226), Expect = e-135
Identities = 228/356 (64%), Positives = 287/356 (80%), Gaps = 1/356 (0%)
Frame = +2
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
MA+ SAIGFEG+EKRLEISF++ G+FADPEG GLR L++ QL EIL PA CTIV SL N
Sbjct: 566 MAMAVSAIGFEGFEKRLEISFFQPGLFADPEGRGLRALTKSQLGEILTPAACTIVSSLKN 745
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
D VDSYVLSESSLFVY YK+IIKTCGTTKLLL+IP ILK A+ L++ VKSV YTRGSFIF
Sbjct: 746 DNVDSYVLSESSLFVYAYKIIIKTCGTTKLLLAIPPILKFAEMLSLNVKSVNYTRGSFIF 925
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVY 180
P AQP+PHRNFSEEVA+L+ YFG LG+G AY++G DK+Q WH+Y+ASA+ + VY
Sbjct: 926 PSAQPYPHRNFSEEVAILDGYFGKLGAGSNAYILGGQDKAQNWHVYSASADSVTQCDNVY 1105
Query: 181 GLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEG 240
LEMCMTGLD+E A VF+K+ ++SAA+MT NSGI KILP S+ICDF+F+PCGYSMN +EG
Sbjct: 1106TLEMCMTGLDREKAQVFYKEQSASAAMMTVNSGIRKILPDSEICDFDFEPCGYSMNSVEG 1285
Query: 241 SAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKL 300
+A+STIHVTPEDGFSYASFE VGYD++ ++L E+V+RVLACF P EFSVA+H+D +
Sbjct: 1286AAVSTIHVTPEDGFSYASFETVGYDFKAVNLNEMVQRVLACFLPTEFSVAVHVDGASKSF 1465
Query: 301 DKFP-LDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDE 355
++ LD++GY ++R E LG GG+V++ F + C SP+S LK CW+E++ +E
Sbjct: 1466EQTCFLDVKGYCREERSHEGLGMGGSVVYQKFGKTSDCGSPRSTLK-CWNEEDEEE 1630
>TC204353 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase,
partial (58%)
Length = 1265
Score = 283 bits (723), Expect = 1e-76
Identities = 137/201 (68%), Positives = 164/201 (81%)
Frame = +3
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
MA+ SAIGFEG+EKRLEISF++ G+FADPEG+GLR L++ QLDEIL PA CTIV SL N
Sbjct: 585 MAMAVSAIGFEGFEKRLEISFFQPGLFADPEGMGLRALAKSQLDEILTPAACTIVSSLRN 764
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
D+VDSYVLSESSLFVY YK+IIKTCGTTKLLL+IP ILK A+ L++ V+SV YTRGSFIF
Sbjct: 765 DHVDSYVLSESSLFVYAYKIIIKTCGTTKLLLAIPPILKFAEMLSLNVRSVNYTRGSFIF 944
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVY 180
P AQP+PHRNFSEEVA+L+ YFG L +G AY++G DKSQ WH+Y+ASA+ + VY
Sbjct: 945 PGAQPYPHRNFSEEVAILDGYFGKLSAGSNAYILGGQDKSQNWHVYSASADSVTPCDNVY 1124
Query: 181 GLEMCMTGLDKESASVFFKDN 201
LEMCMTGLD+E K+N
Sbjct: 1125TLEMCMTGLDREKHRFSTKNN 1187
>TC204355 similar to UP|Q852S8 (Q852S8) S-adenosylmethionine decarboxylase,
partial (42%)
Length = 774
Score = 282 bits (722), Expect = 1e-76
Identities = 136/167 (81%), Positives = 152/167 (90%), Gaps = 1/167 (0%)
Frame = +3
Query: 195 SVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPEDGF 254
SVFFK+NTSSAA MT NSGI KILPQSDI DFEFDPCGYSMNGIEGSAISTIHVTPEDGF
Sbjct: 3 SVFFKENTSSAASMTENSGIRKILPQSDISDFEFDPCGYSMNGIEGSAISTIHVTPEDGF 182
Query: 255 SYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKFPLDIEGYYCDK 314
SYASFEAVGYD+ D++L ELVERVLACF PAEFSVALH+DMHGEKL+KFPLDI+GYYC +
Sbjct: 183 SYASFEAVGYDFNDMALGELVERVLACFCPAEFSVALHIDMHGEKLNKFPLDIKGYYCGE 362
Query: 315 RGTEELGAGGAVMFHSFVR-VDGCASPKSILKCCWSEDENDEEVKGI 360
R TEELG GGAV++ +FV+ DG ASP+SILKCCWSEDEN++EV+ I
Sbjct: 363 RSTEELGVGGAVVYQTFVQGCDGSASPRSILKCCWSEDENEDEVREI 503
>TC204359 similar to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase,
partial (71%)
Length = 835
Score = 275 bits (704), Expect = 2e-74
Identities = 143/279 (51%), Positives = 189/279 (67%), Gaps = 2/279 (0%)
Frame = +2
Query: 55 VGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYT 114
V L N DSY L +S L Y +II TCGTTKL L+ ILK + + + +S T
Sbjct: 2 VSFLKNHTGDSYGLWDSRLLDYACNIIIPTCGTTKLSLANAPILKGTEMRDRSDRSENNT 181
Query: 115 RGSFIFPEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKG 174
RGSFI P A P PHR FS+EVA+L+ Y G LG+ A ++G DK+Q WH+Y+ASA+
Sbjct: 182 RGSFICPRAHPCPHRTFSDEVAILDGYLGELGA*SNANILGGQDKAQNWHVYSASADSVT 361
Query: 175 SSEAVYGLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYS 234
+ VY LEMCMTGLD+E A VF+K+ ++SAA+MT NSGI KILP ++ICD + +PCGYS
Sbjct: 362 QCDNVYTLEMCMTGLDREKAQVFYKEQSASAAMMTVNSGIRKILPDAEICDCDLEPCGYS 541
Query: 235 MNGIEGSAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMD 294
MN +E + +STIHVTPEDG S ASFE VGYD++ ++L ++V+RVLACF P EFSVA+H+D
Sbjct: 542 MNSVERAVVSTIHVTPEDGCSSASFETVGYDFKAVNLNDMVQRVLACFLPTEFSVAVHVD 721
Query: 295 MHGEKLDKF-PLDIEGYYCDKRGTE-ELGAGGAVMFHSF 331
++ + LD++G YCD L GG+ H F
Sbjct: 722 GASDEYEHTRVLDVKG-YCDVETIHLRLAVGGSDDLHRF 835
>TC204358 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase,
partial (61%)
Length = 953
Score = 259 bits (661), Expect = 2e-69
Identities = 122/219 (55%), Positives = 163/219 (73%), Gaps = 1/219 (0%)
Frame = +3
Query: 138 LNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGLEMCMTGLDKESASVF 197
L+ Y L +G AY G DKSQ WH+Y+ASA+ + VY LEMCMTGLD+E A VF
Sbjct: 3 LDGYXXKLSAGSNAYXXGXQDKSQNWHVYSASADSVTPCDNVYTLEMCMTGLDREKAQVF 182
Query: 198 FKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPEDGFSYA 257
+K+ ++SAA+MT NSGI KILP S+ICDF+F+PCGYSMN +EG+A+STIHVTPEDGFSYA
Sbjct: 183 YKEQSASAAIMTVNSGIRKILPDSEICDFDFEPCGYSMNSVEGAAVSTIHVTPEDGFSYA 362
Query: 258 SFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKFP-LDIEGYYCDKRG 316
SFE VGYD++ ++L E+V+RVLACF P EFSVA+H+D + D+ LD++GY ++R
Sbjct: 363 SFETVGYDFKVVNLNEMVKRVLACFLPTEFSVAVHVDGASKLFDQTCFLDVKGYCREERS 542
Query: 317 TEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDE 355
E LG GG++++ F + C SP+S LK CW E++ +E
Sbjct: 543 HEGLGMGGSLVYQKFAKTCDCGSPRSTLK-CWKEEDEEE 656
>TC204356 similar to UP|Q6KC47 (Q6KC47) S-adenosylmethionine decarboxylase
(Fragment), partial (51%)
Length = 795
Score = 194 bits (493), Expect = 5e-50
Identities = 97/111 (87%), Positives = 104/111 (93%)
Frame = +1
Query: 3 LTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDY 62
+ SAIGFEGYEKRLEISF+E GVFADP GLGLR LS+DQLDEIL PA+CTIV SLSNDY
Sbjct: 463 MAGSAIGFEGYEKRLEISFFENGVFADPGGLGLRALSKDQLDEILKPAECTIVASLSNDY 642
Query: 63 VDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRY 113
VDSYVLSESSLFVYPYK+IIKTCGTTKLLLSIPVILKLAD+L+IAVKSVRY
Sbjct: 643 VDSYVLSESSLFVYPYKIIIKTCGTTKLLLSIPVILKLADALDIAVKSVRY 795
>BF009939 weakly similar to GP|18152533|emb S-adenosyl-L-methionine
decarboxylase {Vitis vinifera}, partial (41%)
Length = 385
Score = 160 bits (406), Expect = 6e-40
Identities = 76/126 (60%), Positives = 103/126 (81%)
Frame = +2
Query: 6 SAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDS 65
SAI F+ Y+K LEISF++ VF +P+ L L+ L RDQL++ILNPA+ TI+ SLSN+Y+DS
Sbjct: 8 SAINFKSYKKILEISFFKNNVFTNPKELSLQSLCRDQLNKILNPAEYTIITSLSNNYIDS 187
Query: 66 YVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPEAQP 125
Y+LSES+LF+YPYK+IIKT TTKLLLSIP+ILKL ++L+I++K ++YTR + I P+ Q
Sbjct: 188 YILSESNLFIYPYKIIIKTYKTTKLLLSIPIILKLTNALDISIKCIKYTRKTCIVPKTQS 367
Query: 126 FPHRNF 131
FPH F
Sbjct: 368 FPHAQF 385
>AW132199 similar to GP|19526455|gb S-adenosylmethionine decarboxylase
{Glycine max}, partial (38%)
Length = 442
Score = 157 bits (398), Expect = 5e-39
Identities = 84/147 (57%), Positives = 102/147 (69%)
Frame = +2
Query: 2 ALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSND 61
A+ SAIGFEG EKRLE S++ G F P G G + +IL PA CTI S+ ND
Sbjct: 2 AMAGSAIGFEG*EKRLETSYFPSGTFC*P*GNGYKSSCTVPPYDILTPAACTIASSVRND 181
Query: 62 YVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFP 121
+V SYVLSESSLFVY YK+IIKTCGTT LL +IP ILK A+ L++ SV YTRGS IFP
Sbjct: 182 HVHSYVLSESSLFVYAYKIIIKTCGTT*LLFAIPPILKPAEMLSLGAISVNYTRGSGIFP 361
Query: 122 EAQPFPHRNFSEEVAVLNNYFGNLGSG 148
AQ + HRN S +VA+++ YFG L +G
Sbjct: 362 RAQSYRHRNSSGDVAMIDGYFG*LSAG 442
>CK605936
Length = 707
Score = 145 bits (365), Expect = 4e-35
Identities = 80/177 (45%), Positives = 109/177 (61%), Gaps = 2/177 (1%)
Frame = -1
Query: 183 EMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSA 242
E+CMTGLD++ A VF K+ MT SGI++I+ ICDFEF+PCGYSMNGIEG+A
Sbjct: 707 EICMTGLDRDKARVFHKE--WGGGKMTEMSGISEIVQSHVICDFEFEPCGYSMNGIEGAA 534
Query: 243 ISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDK 302
ST+HVTPE+GFSY S+E G D LV +VL CF P++FSVA+ ++ E
Sbjct: 533 FSTVHVTPENGFSYGSYEVQGLDPGSNGFGALVRKVLKCFCPSDFSVAVTCEVGAEDWAM 354
Query: 303 FPLDIEGYYCDKRGTEELGAG-GAVMFHSF-VRVDGCASPKSILKCCWSEDENDEEV 357
D+EGY C ++L G G +++ ++ R G SPK + C +E E +E V
Sbjct: 353 SDADVEGYCCQNVVKQQLLPGKGCLVYRTYSARGRGRGSPKIAIVKCSNEIEKEELV 183
>BF008687
Length = 436
Score = 132 bits (333), Expect = 2e-31
Identities = 68/146 (46%), Positives = 99/146 (67%), Gaps = 1/146 (0%)
Frame = +2
Query: 211 NSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPEDGFSYASFEAVGYDYEDLS 270
NSGI KILP S+I DF+F+PCGYSM +E +A+ST V+PE+GFSYASF+ VGYD+ ++
Sbjct: 5 NSGIRKILPDSEI*DFDFEPCGYSMISVESAAVSTFLVSPEEGFSYASFDTVGYDF*AVN 184
Query: 271 LTELVERVLACFHPAEFSVALHMDMHGEKLDKFP-LDIEGYYCDKRGTEELGAGGAVMFH 329
L E+V++VLACF P EFSVA+H+D + ++ LD+ GY ++R LG + +H
Sbjct: 185 LNEMVQKVLACFLPTEFSVAVHVDGASKSFEQTCFLDV*GYCREERSLVRLGLVSLLSYH 364
Query: 330 SFVRVDGCASPKSILKCCWSEDENDE 355
+ + SP S CW++D+ D+
Sbjct: 365 -YASILVTVSPIS-TS*CWNDDDGDD 436
>AW100067 similar to GP|21239731|gb| S-adenosylmethionine decarboxylase {x
Citrofortunella mitis}, partial (18%)
Length = 259
Score = 115 bits (287), Expect = 4e-26
Identities = 60/87 (68%), Positives = 67/87 (76%)
Frame = +2
Query: 7 AIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDSY 66
AIGF K L I F G ADPEG+GL L +DQLDEIL PA+CTIV SLSNDYVDSY
Sbjct: 2 AIGFHAMNKTLNIIF*H-GALADPEGIGLPALFKDQLDEILKPAKCTIVASLSNDYVDSY 178
Query: 67 VLSESSLFVYPYKVIIKTCGTTKLLLS 93
VLSE+SLF+YPY +II+T TTKLLLS
Sbjct: 179VLSEASLFIYPYTIIIRTSWTTKLLLS 259
>AW156140 weakly similar to GP|19526455|gb| S-adenosylmethionine
decarboxylase {Glycine max}, partial (16%)
Length = 425
Score = 83.2 bits (204), Expect = 2e-16
Identities = 54/137 (39%), Positives = 75/137 (54%), Gaps = 5/137 (3%)
Frame = +2
Query: 57 SLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRG 116
S+ ND+ S LSES+L+ Y +K +I TCGTTKLLL+IP +LK ++L++ SV YTRG
Sbjct: 8 SVRNDHAYSCGLSESNLYDYAFKSLI*TCGTTKLLLAIPPLLKFTETLSLKAYSVNYTRG 187
Query: 117 SFIFPE-----AQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAE 171
+ A P R+ S A+L + Q G PDK+Q W +Y A+
Sbjct: 188 RSDLSQG*LIFASPLI*RS-SYSRALLRATCCRM----QCSYCGWPDKTQSWRVYCVLAD 352
Query: 172 PKGSSEAVYGLEMCMTG 188
+ Y +EMCMTG
Sbjct: 353 TVSDCDNEYTVEMCMTG 403
>TC204357 similar to UP|Q6KC47 (Q6KC47) S-adenosylmethionine decarboxylase
(Fragment), partial (28%)
Length = 729
Score = 68.6 bits (166), Expect = 4e-12
Identities = 31/39 (79%), Positives = 35/39 (89%)
Frame = +3
Query: 3 LTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRD 41
+ SAIGFEGYEKRLEISF+E GVFADPEG+GLR LS+D
Sbjct: 543 MAGSAIGFEGYEKRLEISFFEHGVFADPEGIGLRALSKD 659
>AI966554 similar to GP|19526455|gb| S-adenosylmethionine decarboxylase
{Glycine max}, partial (17%)
Length = 199
Score = 57.8 bits (138), Expect = 8e-09
Identities = 26/50 (52%), Positives = 35/50 (70%)
Frame = +2
Query: 245 TIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMD 294
+IHVTP++ FSYAS V YD+ ++L E+V+ VL CF P E S A+H D
Sbjct: 2 SIHVTPKNRFSYASLTTVDYDFNAVNLNEMVQTVLGCFLPNEVSAAVHED 151
>AI856221
Length = 124
Score = 36.6 bits (83), Expect = 0.018
Identities = 16/39 (41%), Positives = 26/39 (66%)
Frame = -1
Query: 180 YGLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKIL 218
Y L MCM + +E+A + ++D ++A MT N+GI +IL
Sbjct: 124 YTLVMCMLYMYRETAQMVYRDQPATAPTMTVNAGIREIL 8
>TC209845 similar to PIR|E87912|E87912 protein B0205.8 [imported] -
Caenorhabditis elegans {Caenorhabditis elegans;} ,
partial (16%)
Length = 720
Score = 29.6 bits (65), Expect = 2.2
Identities = 14/46 (30%), Positives = 24/46 (51%)
Frame = -3
Query: 238 IEGSAISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFH 283
++ S+I +TP + FS A+ V L L+E +++CFH
Sbjct: 457 VDSDFCSSILITPWNSFSIATPTIVATTRPARELASLLETIISCFH 320
>TC215479
Length = 1561
Score = 29.3 bits (64), Expect = 2.9
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 14/100 (14%)
Frame = +3
Query: 211 NSGINKILPQSDIC------------DFEFDPCGYSMNGIEGSAISTI--HVTPEDGFSY 256
N + ++LP S + D F P + + G S I HV+P DG+ +
Sbjct: 120 NYSVRRVLPMSRVVLRSYATVTAKPNDIPFQPKLANSVNLIGHVQSPIQFHVSPNDGYVW 299
Query: 257 ASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMH 296
AS D DLS + V H A+F + L+ +H
Sbjct: 300 ASTVITRQDSSDLSFSIPVIFEGDLAHTAKFHLNLNDCIH 419
>TC218005 similar to UP|Q8RVB2 (Q8RVB2) SPY protein, partial (19%)
Length = 1395
Score = 28.9 bits (63), Expect = 3.8
Identities = 21/67 (31%), Positives = 31/67 (45%)
Frame = +1
Query: 31 EGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKL 90
E LGL L D L IL L++D++ +Y L + SL +PY TC L
Sbjct: 70 EKLGLEPLRVDLLPLIL----------LNHDHMQAYSLMDISLDTFPYAGTTTTC--ESL 213
Query: 91 LLSIPVI 97
+ +P +
Sbjct: 214 YMGVPCV 234
>TC232613 similar to UP|Q8LEH1 (Q8LEH1) Ripening-related protein-like,
partial (10%)
Length = 861
Score = 27.7 bits (60), Expect = 8.4
Identities = 17/48 (35%), Positives = 22/48 (45%)
Frame = +1
Query: 75 VYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPE 122
+YPY IK T LSI V LK A S N + + ++G E
Sbjct: 196 LYPYASQIKRNSVTLTKLSIHVALKAAKSANSTLTKLSNSKGKLTHGE 339
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,026,676
Number of Sequences: 63676
Number of extensions: 216340
Number of successful extensions: 910
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 892
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 904
length of query: 360
length of database: 12,639,632
effective HSP length: 98
effective length of query: 262
effective length of database: 6,399,384
effective search space: 1676638608
effective search space used: 1676638608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0215b.11


