

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
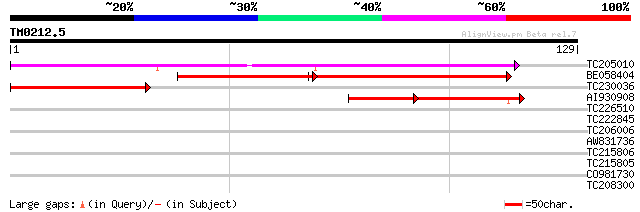
Query= TM0212.5
(129 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205010 UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate reductas... 105 6e-24
BE058404 similar to GP|18252504|gb adenosine 5'-phosphosulfate r... 69 3e-18
TC230036 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosul... 49 6e-07
AI930908 similar to GP|18252504|gb| adenosine 5'-phosphosulfate ... 31 3e-04
TC226510 similar to GB|BAC55689.1|27817924|AP003940 contains EST... 27 3.4
TC222845 23 5.3
TC206006 weakly similar to UP|Q9LKW3 (Q9LKW3) Dehydration-induce... 26 5.9
AW831736 25 7.7
TC215806 similar to UP|Q9FGN2 (Q9FGN2) Gb|AAB68038.1, partial (54%) 25 7.7
TC215805 25 7.7
CO981730 25 7.7
TC208300 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase,... 25 7.7
>TC205010 UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate reductase, complete
Length = 1921
Score = 105 bits (262), Expect = 6e-24
Identities = 66/143 (46%), Positives = 78/143 (54%), Gaps = 27/143 (18%)
Frame = +2
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 647 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGIGSLVKWNP 826
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 827 VANVNGLDIWSFLRT-MDVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 1003
Query: 94 DCGLHKGNVKQDVGAQLNGNGAA 116
+CGLHKGN+K + AQLNGNGA+
Sbjct: 1004ECGLHKGNIKHEDAAQLNGNGAS 1072
>BE058404 similar to GP|18252504|gb adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (28%)
Length = 433
Score = 68.9 bits (167), Expect(2) = 3e-18
Identities = 31/46 (67%), Positives = 33/46 (71%)
Frame = +1
Query: 69 PLEALVLREQHEREGNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNG 114
P VL QHEREG WWEDAKAK+CGLHKGNVKQ +NGNG
Sbjct: 112 PCTRPVLPGQHEREGRWWWEDAKAKECGLHKGNVKQQKEEDVNGNG 249
Score = 38.1 bits (87), Expect(2) = 3e-18
Identities = 19/32 (59%), Positives = 25/32 (77%)
Frame = +2
Query: 39 LVAISKAIIYGISLGP*MCL*IH*IRKDTFPL 70
L+ + +A+ YG SLGP*MCL*I ++KD FPL
Sbjct: 8 LLQM*RAMTYGTSLGP*MCL*IPCMQKDMFPL 103
>TC230036 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate
reductase, partial (41%)
Length = 750
Score = 48.9 bits (115), Expect = 6e-07
Identities = 24/32 (75%), Positives = 25/32 (78%)
Frame = +3
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT 32
CCRVRKVR LRR LKG+ I GQ KDQSPGT
Sbjct: 612 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGT 707
>AI930908 similar to GP|18252504|gb| adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (16%)
Length = 404
Score = 30.8 bits (68), Expect(2) = 3e-04
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 3/28 (10%)
Frame = +2
Query: 93 KDCGLHKGNVKQDVGAQLNG---NGAAM 117
++CGLHKGN+K ++ NG NG+A+
Sbjct: 257 QECGLHKGNIKHEICHLWNGPPSNGSAL 340
Score = 28.5 bits (62), Expect(2) = 3e-04
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +1
Query: 78 QHEREGNRWWEDAKAK 93
QHER+ WWEDAK +
Sbjct: 214 QHERKEGVWWEDAKPR 261
>TC226510 similar to GB|BAC55689.1|27817924|AP003940 contains ESTs
AU077521(C52420),AU077522(C52420) snRNP core Sm protein
Sm-X5-like protein {Oryza sativa (japonica
cultivar-group);} , complete
Length = 562
Score = 26.6 bits (57), Expect = 3.4
Identities = 14/31 (45%), Positives = 18/31 (57%), Gaps = 7/31 (22%)
Frame = -2
Query: 66 DTFPLEAL--VLREQH-----EREGNRWWED 89
D FP + V EQH ER+G+RWWE+
Sbjct: 132 DYFPPHQILEVREEQHVCAEGERKGSRWWEN 40
>TC222845
Length = 643
Score = 23.5 bits (49), Expect(2) = 5.3
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = -1
Query: 31 GTNLVKWNLVAISKA 45
GTNL KW+L ++ KA
Sbjct: 265 GTNLRKWSLKSLEKA 221
Score = 20.8 bits (42), Expect(2) = 5.3
Identities = 7/15 (46%), Positives = 8/15 (52%)
Frame = -2
Query: 74 VLREQHEREGNRWWE 88
+ R H RE R WE
Sbjct: 219 IQRRSHHRESRRRWE 175
>TC206006 weakly similar to UP|Q9LKW3 (Q9LKW3) Dehydration-induced protein
ERD15, partial (35%)
Length = 1057
Score = 25.8 bits (55), Expect = 5.9
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = -2
Query: 110 LNGNGAAMQVHCHCRIHLK 128
LN N A +VH HC I LK
Sbjct: 1023 LNANHKA*KVHSHCHIELK 967
>AW831736
Length = 396
Score = 25.4 bits (54), Expect = 7.7
Identities = 11/26 (42%), Positives = 18/26 (68%)
Frame = +3
Query: 27 DQSPGTNLVKWNLVAISKAIIYGISL 52
D+ T L+ WNL ++S+ I+ GIS+
Sbjct: 186 DKGMQTCLIFWNLCSVSRKIM*GISV 263
>TC215806 similar to UP|Q9FGN2 (Q9FGN2) Gb|AAB68038.1, partial (54%)
Length = 732
Score = 25.4 bits (54), Expect = 7.7
Identities = 12/42 (28%), Positives = 18/42 (42%), Gaps = 1/42 (2%)
Frame = +1
Query: 86 WWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQVHC-HCRIH 126
WW CG+H G V+Q + + + + C HC H
Sbjct: 256 WWM------CGIHTGQVRQSLQKKYHLKNSPCNACCVHCCFH 363
>TC215805
Length = 783
Score = 25.4 bits (54), Expect = 7.7
Identities = 12/42 (28%), Positives = 18/42 (42%), Gaps = 1/42 (2%)
Frame = +2
Query: 86 WWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQVHC-HCRIH 126
WW CG+H G V+Q + + + + C HC H
Sbjct: 545 WWM------CGIHTGQVRQSLQKKYHLKNSPCNACCVHCCFH 652
>CO981730
Length = 861
Score = 25.4 bits (54), Expect = 7.7
Identities = 12/36 (33%), Positives = 21/36 (58%)
Frame = -2
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQ 118
GN W + ++CG +K NVK++ +L A++Q
Sbjct: 524 GNAW---SVHEECGANKANVKEETSWKLAQREASVQ 426
>TC208300 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, partial
(31%)
Length = 1014
Score = 25.4 bits (54), Expect = 7.7
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +3
Query: 76 REQHEREGNRWWEDAKAKDCGLHKGNVKQD 105
R+Q E G+ WE+ +A DC +G +++D
Sbjct: 282 RDQGEASGSGAWEEGEAGDC---RGYLRRD 362
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.344 0.153 0.534
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,102,571
Number of Sequences: 63676
Number of extensions: 69851
Number of successful extensions: 886
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 877
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 884
length of query: 129
length of database: 12,639,632
effective HSP length: 86
effective length of query: 43
effective length of database: 7,163,496
effective search space: 308030328
effective search space used: 308030328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0212.5


