

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
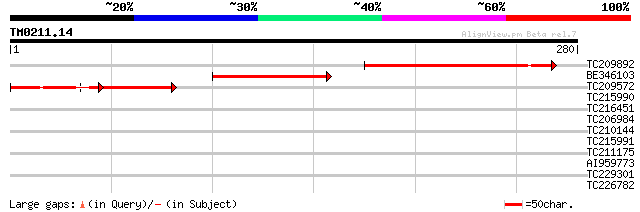
Query= TM0211.14
(280 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209892 UP|Q7PHQ2 (Q7PHQ2) ENSANGP00000022785 (Fragment), parti... 103 7e-23
BE346103 77 9e-15
TC209572 48 6e-06
TC215990 32 0.42
TC216451 similar to UP|Q93XR7 (Q93XR7) Fructose-6-phosphate 2-ki... 30 0.93
TC206984 similar to UP|Q9SSR5 (Q9SSR5) F6D8.15 protein, partial ... 30 0.93
TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, part... 30 0.93
TC215991 29 2.1
TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell... 28 6.0
AI959773 homologue to GP|4467130|emb| glycosyltransferase like p... 27 7.9
TC229301 similar to UP|Q8YLU6 (Q8YLU6) Alr5200 protein, partial ... 27 7.9
TC226782 homologue to UP|ERF3_ARATH (O80339) Ethylene responsive... 27 7.9
>TC209892 UP|Q7PHQ2 (Q7PHQ2) ENSANGP00000022785 (Fragment), partial (14%)
Length = 582
Score = 103 bits (258), Expect = 7e-23
Identities = 49/95 (51%), Positives = 65/95 (67%)
Frame = +1
Query: 176 GLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPE 235
GLT R+FLM+W+KWTV+QFE LNN GN VME+G GG YSL +HH EEE+ ++GL+ E
Sbjct: 1 GLTLRVFLMRWYKWTVQQFESLNNMGNGNMLVMEKGFGGRYSLLMHHGEEELRRFGLTDE 180
Query: 236 MVADQKWRAGASRGAWNDQCSWYLDGFFDRLAEES 270
M+ DQ+W A N C ++ FF L+EE+
Sbjct: 181 MLIDQEWHKFAKPADLNYDCP-MVNSFFPHLSEEA 282
>BE346103
Length = 184
Score = 77.0 bits (188), Expect = 9e-15
Identities = 35/59 (59%), Positives = 44/59 (74%)
Frame = +2
Query: 101 REQDFGNFQVEERMKIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNR 159
REQD+GNFQ + M++ K R+R+GRFFYR P GESA DVYDRI+G E L DI++ R
Sbjct: 2 REQDWGNFQHRDEMRVEKALRQRYGRFFYRLPYGESAVDVYDRITGCIETLRTDINIGR 178
>TC209572
Length = 370
Score = 47.8 bits (112), Expect = 6e-06
Identities = 25/47 (53%), Positives = 30/47 (63%)
Frame = -2
Query: 36 TPQGISQARRAGNRLRRVMAADGCSPDWRVNFYVSPYARTRSTLREV 82
T QG++ A R RRV+ +D SPDWRV F VS YARTRS E+
Sbjct: 255 TAQGMA*ALRPDKHFRRVIGSDDYSPDWRV*F*VSTYARTRSMFYEL 115
Score = 41.2 bits (95), Expect = 5e-04
Identities = 25/46 (54%), Positives = 31/46 (67%)
Frame = -3
Query: 1 MGVLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRA 46
+GV ++ ILM HGE QGN DT+ YTTTPD+SI SQ R+A
Sbjct: 359 VGVFLEKRILM*HGE-QGN*DTTTYTTTPDYSI------YSQQRKA 243
>TC215990
Length = 884
Score = 31.6 bits (70), Expect = 0.42
Identities = 21/80 (26%), Positives = 34/80 (42%)
Frame = +2
Query: 200 FGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKWRAGASRGAWNDQCSWYL 259
+GN F V+ +H T E +Q A++KWRA G+ +DQ ++
Sbjct: 179 YGNDSFYVL---------FRLHQTLYERIQSAKVNSSSAEKKWRASNDTGS-SDQYGRFM 328
Query: 260 DGFFDRLAEESDDEEASSSC 279
D ++ L SD + C
Sbjct: 329 DALYNLLDGSSDSTKFEDEC 388
>TC216451 similar to UP|Q93XR7 (Q93XR7) Fructose-6-phosphate
2-kinase/fructose-2,6-bisphosphatase, partial (62%)
Length = 1595
Score = 30.4 bits (67), Expect = 0.93
Identities = 38/146 (26%), Positives = 63/146 (43%), Gaps = 6/146 (4%)
Frame = +1
Query: 5 PKRIILMRHGESQGNL------DTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADG 58
P+ I+L RHGESQ N+ DT+ +S +L A+ G RL+ AA
Sbjct: 802 PRPILLTRHGESQDNVRGRIGGDTAISEAGELYSKKL-------AKFVGKRLKSERAA-- 954
Query: 59 CSPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIK 118
+ + S RT T + F K + + E + ++++ M
Sbjct: 955 -------SIWTSTLQRTILTATPI-VGFPKIQWRALDEINAGVCDGMTYAEIKKNMPEEY 1110
Query: 119 QTRERFGRFFYRFPEGESAADVYDRI 144
++R++ + YR+P GES DV R+
Sbjct: 1111ESRKK-DKLRYRYPRGESYLDVIQRL 1185
>TC206984 similar to UP|Q9SSR5 (Q9SSR5) F6D8.15 protein, partial (51%)
Length = 744
Score = 30.4 bits (67), Expect = 0.93
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 6/48 (12%)
Frame = -1
Query: 152 WRDIDLNRLHHDPSNDLNLVIVSHGLT--SRIF----LMKWFKWTVEQ 193
W++I++ +L H D++ V+ HGLT S +F L+ W W E+
Sbjct: 204 WKEIEVLKLSH*A*VDISHVVSKHGLTLQSMVFCPLVLLLWLSWKQEK 61
>TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, partial (54%)
Length = 1167
Score = 30.4 bits (67), Expect = 0.93
Identities = 15/44 (34%), Positives = 22/44 (49%)
Frame = +2
Query: 148 FEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFLMKWFKWTV 191
+ +W D N HDP +L L+I SH + IF M + +V
Sbjct: 923 YSGMWN*CDWNATSHDPWENLFLLIRSHQILFYIFFMLFVSTSV 1054
>TC215991
Length = 1356
Score = 29.3 bits (64), Expect = 2.1
Identities = 16/62 (25%), Positives = 28/62 (44%)
Frame = +1
Query: 218 LAVHHTEEEMLQWGLSPEMVADQKWRAGASRGAWNDQCSWYLDGFFDRLAEESDDEEASS 277
L +H T E +Q AD+KW+A + + DQ +++ + L SD+ +
Sbjct: 232 LRLHQTLYERIQSAKINSSSADRKWKASSDTSS-TDQYDRFMNALYSLLDGSSDNTKFED 408
Query: 278 SC 279
C
Sbjct: 409 DC 414
>TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell wall-plasma
membrane linker protein, partial (3%)
Length = 609
Score = 27.7 bits (60), Expect = 6.0
Identities = 20/71 (28%), Positives = 30/71 (42%)
Frame = -1
Query: 44 RRAGNRLRRVMAADGCSPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQ 103
R +G R RR SPDWR + + R RS GRS ++ ++G R R
Sbjct: 498 RNSGRRFRRRRLCHWRSPDWRTH---RSHRRRRS---RWGRSRRRRNIVGWAGSRRNRRW 337
Query: 104 DFGNFQVEERM 114
++ R+
Sbjct: 336 GLSGRRLRRRV 304
>AI959773 homologue to GP|4467130|emb| glycosyltransferase like protein
{Arabidopsis thaliana}, partial (17%)
Length = 455
Score = 27.3 bits (59), Expect = 7.9
Identities = 20/61 (32%), Positives = 30/61 (48%), Gaps = 1/61 (1%)
Frame = +1
Query: 113 RMKIIKQTRERFGRFFYRFPEGESAADVYDRISGFFE-ALWRDIDLNRLHHDPSNDLNLV 171
R+ I++ E FF F E+AA + ++ + W D + HDPS+ LNLV
Sbjct: 58 RVIILEDDMEIAPDFFDYF---EAAASLLEKDKSIMAVSSWNDNGQKQFVHDPSSALNLV 228
Query: 172 I 172
I
Sbjct: 229 I 231
>TC229301 similar to UP|Q8YLU6 (Q8YLU6) Alr5200 protein, partial (11%)
Length = 517
Score = 27.3 bits (59), Expect = 7.9
Identities = 18/49 (36%), Positives = 26/49 (52%)
Frame = +2
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRV 53
PK++ L+RHG S N + S + D S+ LT G QA+R L +
Sbjct: 263 PKKVTLLRHGLSTWNSE-SRIQGSSDLSV-LTEVGEEQAKRCKKALENI 403
>TC226782 homologue to UP|ERF3_ARATH (O80339) Ethylene responsive element
binding factor 3 (AtERF3), partial (48%)
Length = 1194
Score = 27.3 bits (59), Expect = 7.9
Identities = 15/29 (51%), Positives = 16/29 (54%)
Frame = +1
Query: 71 PYARTRSTLREVGRSFSKKRVIGVREESR 99
P R RS GR FSK RVIG E+ R
Sbjct: 355 PRRRWRSPNPVQGRFFSKNRVIGASEKDR 441
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,866,539
Number of Sequences: 63676
Number of extensions: 174220
Number of successful extensions: 830
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 827
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 829
length of query: 280
length of database: 12,639,632
effective HSP length: 96
effective length of query: 184
effective length of database: 6,526,736
effective search space: 1200919424
effective search space used: 1200919424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0211.14


