

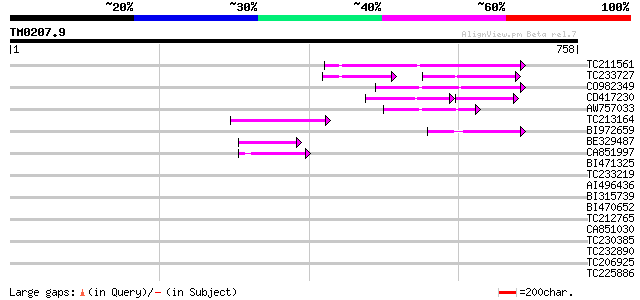
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.9
(758 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 125 5e-29
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 67 3e-17
CO982349 82 9e-16
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 59 3e-15
AW757033 68 1e-11
TC213164 64 2e-10
BI972659 48 1e-05
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 46 5e-05
CA851997 43 6e-04
BI471325 42 0.001
TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance... 40 0.003
AI496436 39 0.009
BI315739 38 0.019
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 36 0.073
TC212765 33 0.61
CA851030 32 1.0
TC230385 weakly similar to UP|Q9M019 (Q9M019) Receptor like prot... 31 1.8
TC232890 30 3.1
TC206925 homologue to UP|Q39844 (Q39844) Small GTP-binding prote... 30 3.1
TC225886 similar to UP|Q9LRE9 (Q9LRE9) Cytosolic aldehyde dehydr... 29 6.8
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 125 bits (315), Expect = 5e-29
Identities = 85/268 (31%), Positives = 127/268 (46%)
Frame = +1
Query: 422 NALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMD 481
+AL+A E K+ C + K+D KAYD V W FL + +M F W+ +I +
Sbjct: 7 SALIANEVIDEAKRSNKSC---LVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEE 177
Query: 482 YVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIK 541
V + V++NG+P F P RGLRQGDPL+P+LF I E + L++R + +
Sbjct: 178 CVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYL 357
Query: 542 IARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRN 601
+ N IS L +ADD + F A +E +K IL ++ V INF KS
Sbjct: 358 VGAN--GVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVF-G 528
Query: 602 VPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSR 661
V + E L + YLG+P + ++ + + +KL K +S
Sbjct: 529 VTDQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISF 708
Query: 662 AGREVLIKAVAQEIPAYIMSCFILPDNL 689
GR LIK+V IP Y S F +P ++
Sbjct: 709 GGRVTLIKSVLTSIPIYFFSFFRVPQSV 792
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 67.0 bits (162), Expect(2) = 3e-17
Identities = 36/99 (36%), Positives = 57/99 (57%)
Frame = -3
Query: 419 VTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSF 478
+ D V E + + KK++G N +ALK+D+ KA+D ++ FL S L K G+ + ++
Sbjct: 834 IKDCTCVTSEVINMLDKKVFGGN--LALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNW 661
Query: 479 IMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLF 517
I + + + +NG P F RG+RQGDPLSP L+
Sbjct: 660 IRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 40.4 bits (93), Expect(2) = 3e-17
Identities = 35/132 (26%), Positives = 55/132 (41%)
Frame = -2
Query: 552 SHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELK 611
SH++ ADD +IF + T + Y + QVI+ KS + + ++P +
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKI-FIGSIPGQRLRVIS 275
Query: 612 MLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAV 671
LL D Y G+P GK + V ++ KL K LS GR L+ +V
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 672 AQEIPAYIMSCF 683
+ Y S +
Sbjct: 94 IHGMLLYSFSVY 59
>CO982349
Length = 795
Score = 82.0 bits (201), Expect = 9e-16
Identities = 58/200 (29%), Positives = 88/200 (44%)
Frame = +3
Query: 490 VMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAP 549
+++N +P F P RGLRQGDPL+P LF I E + L++ + + + +N
Sbjct: 105 ILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRFNSFLVGKNKEP- 281
Query: 550 VISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNE 609
+S L +ADD + F AT E +K IL ++ INF +S P + E
Sbjct: 282 -VSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGAIWK-PDHWCKE 455
Query: 610 LKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIK 669
L + YLG+P +I + + KL K+ +S GR LI
Sbjct: 456 AAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHISLGGRVTLIN 635
Query: 670 AVAQEIPAYIMSCFILPDNL 689
A+ +P Y S F P +
Sbjct: 636 AILTALPIYFFSFFRAPSKV 695
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 58.9 bits (141), Expect(2) = 3e-15
Identities = 35/119 (29%), Positives = 57/119 (47%)
Frame = -3
Query: 476 VSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGAS 535
V + ++T ++ NG F P G+RQ DP++PYLF++C E S LI +
Sbjct: 686 VQLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKK 507
Query: 536 HLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKS 594
I++ N +ISHL F DD ++F A ++ + L + +N DK+
Sbjct: 506 L*RSIQV--NRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
Score = 41.6 bits (96), Expect(2) = 3e-15
Identities = 25/84 (29%), Positives = 43/84 (50%)
Frame = -2
Query: 597 SYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKE 656
S+S+NV ++ LG+++ D+ +YLG+P + F F+ ++ K+ K
Sbjct: 327 SFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKS 148
Query: 657 PSLSRAGREVLIKAVAQEIPAYIM 680
LS GR L K V +P++IM
Sbjct: 147 NLLSMVGRLTLTK*VV*ALPSHIM 76
>AW757033
Length = 441
Score = 68.2 bits (165), Expect = 1e-11
Identities = 42/130 (32%), Positives = 62/130 (47%)
Frame = -2
Query: 500 FDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADD 559
F P RGLRQGDPL+P LF I E + L++ + +H + + +S L +ADD
Sbjct: 431 FSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKP--VSILQYADD 258
Query: 560 NVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAM 619
+ F AT E +K IL ++ INF KS + +VP + E + +
Sbjct: 257 TIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFG-AISVPD*WRKEAAEFMNCSLL 81
Query: 620 DNYDRYLGLP 629
YLG+P
Sbjct: 80 SLPFSYLGIP 51
>TC213164
Length = 446
Score = 63.9 bits (154), Expect = 2e-10
Identities = 41/133 (30%), Positives = 63/133 (46%)
Frame = +3
Query: 296 ILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPG 355
+L F ++VR ++ K+ G DGL+ F ++FW I+ D+ F V
Sbjct: 15 LLVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLK 194
Query: 356 TIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVP 415
L LIPK+ ++PISL +KI+ L+ + K VLP II Q+ F+
Sbjct: 195 RSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFME 374
Query: 416 GRLVTDNALVAYE 428
GR N ++A E
Sbjct: 375 GRHTLHNVVIANE 413
>BI972659
Length = 453
Score = 48.1 bits (113), Expect = 1e-05
Identities = 31/131 (23%), Positives = 56/131 (42%)
Frame = +1
Query: 559 DNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKA 618
DN+I ++ V G+ Y K ++ F + + ++ LL +
Sbjct: 19 DNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPN-----------WAHDAAQLLNCRQ 165
Query: 619 MDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAY 678
+D YLG+P + S ++ + + KL + LS GR LIK+V +P Y
Sbjct: 166 LDIPFHYLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIY 345
Query: 679 IMSCFILPDNL 689
++S F +P +
Sbjct: 346 LLSFFKIPQRI 378
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 46.2 bits (108), Expect = 5e-05
Identities = 25/85 (29%), Positives = 40/85 (46%)
Frame = -2
Query: 306 VREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLI 365
++ ++Q K+ G DGL+ F + FW + D+ F + P + + LI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 366 PKIKKSVHATHFRPISLCNVPFKII 390
PK+K FRPISL +KI+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIV 121
>CA851997
Length = 636
Score = 42.7 bits (99), Expect = 6e-04
Identities = 33/98 (33%), Positives = 46/98 (46%), Gaps = 2/98 (2%)
Frame = +1
Query: 307 REPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIP 366
R PL Q+ P DG + FY +FW + G+DV C L P ++ T + +I
Sbjct: 292 RRPLLQILP------DGFNLGFYHRFWGMCGEDVFQACCMWLAEGAFPSSVNDTTIAIIL 453
Query: 367 KIKKSVHATHFRPISLCNVPFKI--IT*TLANKLKIVL 402
+I S LCN FKI ++ LA +LK VL
Sbjct: 454 EI**S*RYERS*TNLLCNGGFKILFLSEVLAKRLKNVL 567
>BI471325
Length = 421
Score = 41.6 bits (96), Expect = 0.001
Identities = 23/77 (29%), Positives = 41/77 (52%)
Frame = +2
Query: 540 IKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYS 599
+K+ R ++ ++SHLL D +F R +EA +K IL ++ V+N KS + +S
Sbjct: 17 VKVHRGIT--ILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEICFS 190
Query: 600 RNVPSLYFNELKMLLGV 616
RN + + + LG+
Sbjct: 191 RNTSQIARDSIFFHLGM 241
>TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance
protein-like protein (Fragment), partial (74%)
Length = 781
Score = 40.4 bits (93), Expect = 0.003
Identities = 32/105 (30%), Positives = 41/105 (38%)
Frame = +3
Query: 462 LRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICG 521
L ALEK G V ++ I D V V G F GL QG LSPYLF +
Sbjct: 3 LWKALEKKGVRVAYIRAIQDMYDRVSTSVRTQGGESDDFPITIGLHQGSTLSPYLFTLIL 182
Query: 522 EVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARA 566
+V + IQ + +LFADD V+ +
Sbjct: 183 DVLTEQIQEIVSRC------------------MLFADDIVLLGES 263
>AI496436
Length = 414
Score = 38.9 bits (89), Expect = 0.009
Identities = 20/65 (30%), Positives = 34/65 (51%)
Frame = +1
Query: 625 YLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFI 684
YLG+P + ++ V + +KL K+ +S GR LI +V +P Y++S F
Sbjct: 145 YLGIPIGANSRHSDVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFR 324
Query: 685 LPDNL 689
+PD +
Sbjct: 325 IPDKV 339
>BI315739
Length = 442
Score = 37.7 bits (86), Expect = 0.019
Identities = 19/35 (54%), Positives = 22/35 (62%)
Frame = +3
Query: 511 PLSPYLFLICGEVFSALIQRKIGASHLSGIKIARN 545
PLSPYLF IC E + LIQ K+ IKI+RN
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRN 152
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 35.8 bits (81), Expect = 0.073
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +1
Query: 483 VTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPY 515
+++ +++NG+P F P RGLRQG P P+
Sbjct: 10 LSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
>TC212765
Length = 637
Score = 32.7 bits (73), Expect = 0.61
Identities = 32/142 (22%), Positives = 60/142 (41%), Gaps = 2/142 (1%)
Frame = +3
Query: 514 PYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASC 573
PYLF++C E + I I ++ + ++ +L+ DD +F A + A
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNR--LVPYLMLVDDIFLFINAIGD*AYL 188
Query: 574 VKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIG 633
L + + + +N DKS + S + + + + L + + +YL L I G
Sbjct: 189 ASLTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHG 368
Query: 634 KSKTQIFRFVK--ERDWKKLKG 653
+SK F V+ W + +G
Sbjct: 369 RSKIVDFLLVERIHNPWPRGRG 434
>CA851030
Length = 358
Score = 32.0 bits (71), Expect = 1.0
Identities = 16/42 (38%), Positives = 23/42 (54%)
Frame = +2
Query: 195 RHNLIHFLFRKKSRGASDPRHLGFNMEIRIQNSFIRKRPSGR 236
R IHFL +++S G+ D G +IQ+SFI K G+
Sbjct: 143 RRAWIHFLSKRRSIGSKDQEQFGSRKGTKIQSSFIGKLYRGK 268
>TC230385 weakly similar to UP|Q9M019 (Q9M019) Receptor like protein kinase,
partial (23%)
Length = 699
Score = 31.2 bits (69), Expect = 1.8
Identities = 16/38 (42%), Positives = 22/38 (57%)
Frame = +2
Query: 169 LMLKINFRNCSRGSRQRRWWPKLTWLRHNLIHFLFRKK 206
L+++I+F S R W L WL +LIHF+ RKK
Sbjct: 242 LLIRISFSWWSGLLRSTTWVKFLRWLILSLIHFMMRKK 355
>TC232890
Length = 435
Score = 30.4 bits (67), Expect = 3.1
Identities = 10/29 (34%), Positives = 23/29 (78%)
Frame = +1
Query: 582 DKVYVQVINFDKSMLSYSRNVPSLYFNEL 610
D+ +++NF+KSM+ Y+R +P+++F+ +
Sbjct: 142 DEKIKELLNFNKSMIYYARVIPTVHFSSM 228
>TC206925 homologue to UP|Q39844 (Q39844) Small GTP-binding protein
(Fragment), partial (95%)
Length = 1111
Score = 30.4 bits (67), Expect = 3.1
Identities = 32/134 (23%), Positives = 57/134 (41%), Gaps = 14/134 (10%)
Frame = +1
Query: 582 DKVYVQVINFDKSMLSYSRNVPSLYFNELK-----MLLGVKAMDNYDRYLGLPTIIGKSK 636
DK V ++ +KS L R VP+ E L A++ + T++
Sbjct: 610 DKNIVIILIGNKSDLENQRQVPTEDAKEFAEKEGLFFLETSALEATNVETAFMTVL---- 777
Query: 637 TQIFRFVKERDWKKLKGR-KEPSLSRAGREVLIKAVAQEIPAYIMSC--------FILPD 687
T+IF + +++ + + S S +G+++++ AQEIP M C FI+
Sbjct: 778 TEIFNIINKKNLAASDNQGNDNSASLSGKKIIVPGPAQEIPKRSMCCQ*SQLYL*FIICL 957
Query: 688 NLCADSTGVAMQPV 701
NL V + P+
Sbjct: 958 NLLDFHGSVFLSPI 999
>TC225886 similar to UP|Q9LRE9 (Q9LRE9) Cytosolic aldehyde dehydrogenase,
partial (70%)
Length = 1322
Score = 29.3 bits (64), Expect = 6.8
Identities = 15/43 (34%), Positives = 24/43 (54%)
Frame = +1
Query: 354 PGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLAN 396
P TI+P L+ K VH+ HF I +C+ +K +*T+ +
Sbjct: 1063 PFTILPGFECLLHGSWKMVHSGHFLSICICHEAWKFSS*TVVS 1191
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.334 0.146 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,942,995
Number of Sequences: 63676
Number of extensions: 479957
Number of successful extensions: 3920
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 3846
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3914
length of query: 758
length of database: 12,639,632
effective HSP length: 104
effective length of query: 654
effective length of database: 6,017,328
effective search space: 3935332512
effective search space used: 3935332512
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0207.9


