

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.16
(596 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
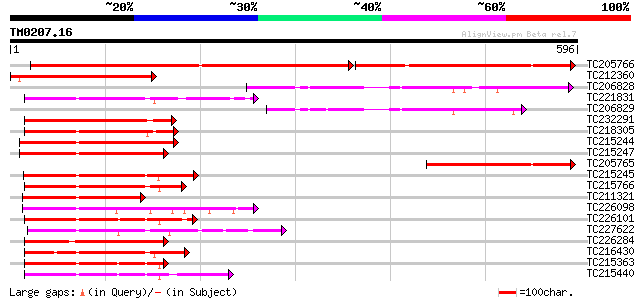
Score E
Sequences producing significant alignments: (bits) Value
TC205766 similar to UP|Q9MAN7 (Q9MAN7) CDS, partial (27%) 478 e-135
TC212360 similar to UP|O81790 (O81790) NAM / CUC2-like protein, ... 288 6e-78
TC206828 similar to UP|Q7VJ29 (Q7VJ29) Biotin operon repressor B... 218 6e-57
TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, com... 180 1e-45
TC206829 UP|Q93ZN0 (Q93ZN0) AT4g35580/F8D20_90, partial (5%) 178 5e-45
TC232291 similar to UP|O81790 (O81790) NAM / CUC2-like protein, ... 174 1e-43
TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like p... 171 8e-43
TC215244 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 160 1e-39
TC215247 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 159 3e-39
TC205765 159 3e-39
TC215245 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 155 4e-38
TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, ... 155 5e-38
TC211321 homologue to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2... 155 6e-38
TC226098 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (45%) 155 6e-38
TC226101 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, part... 154 8e-38
TC227622 similar to UP|Q7XJT9 (Q7XJT9) NAM (No apical meristem)-... 154 1e-37
TC226284 similar to UP|Q9FY93 (Q9FY93) NAM-like protein (AT5g131... 152 3e-37
TC216430 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1... 152 4e-37
TC215363 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, pa... 151 9e-37
TC215440 homologue to UP|Q7Y1A6 (Q7Y1A6) NAC-domain protein 18, ... 151 9e-37
>TC205766 similar to UP|Q9MAN7 (Q9MAN7) CDS, partial (27%)
Length = 2101
Score = 478 bits (1229), Expect = e-135
Identities = 232/341 (68%), Positives = 275/341 (80%), Gaps = 2/341 (0%)
Frame = +1
Query: 23 RPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWFFFCPQDR 82
RP+DEE++++YLRQKINGNG +VWVIREIDVCKWEPWD+P LSV++ +DPEWFFFCPQDR
Sbjct: 1 RPTDEELVNYYLRQKINGNGRQVWVIREIDVCKWEPWDMPGLSVVQTKDPEWFFFCPQDR 180
Query: 83 KYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYR 142
KYPNG+RLNRAT GYWKATGKDRKIKSG +LIGMKKTLVFY GRAPKG RTNWVMHEYR
Sbjct: 181 KYPNGHRLNRATNNGYWKATGKDRKIKSGKNLIGMKKTLVFYTGRAPKGNRTNWVMHEYR 360
Query: 143 PTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKYSPEEVQSDTPL 202
PTL+ELDGTNPGQNPYV+CRLFKK DES+EVS EAE TASTP+AA YS EE+QSD L
Sbjct: 361 PTLKELDGTNPGQNPYVLCRLFKKHDESLEVSHCDEAEPTASTPVAAYYSTEEIQSD--L 534
Query: 203 PLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYDAQNAQPQNVKLAAEENQ 262
+VA S S +TEDDKHQ++IP SEE SN++TPVDR +D YDA +AQ Q AEE Q
Sbjct: 535 AVVAGSPSQVTEDDKHQSMIPAHSEEAISNVVTPVDRRTDGYDACDAQNQIELPTAEEFQ 714
Query: 263 PLNLDM-YNDLKNDIFDDKLFSPALVHLPPVFDYQASNEPDDRSGLQYGTNETAISDLFD 321
PLN D+ Y+D ++++ D KLFSP L H+ P F YQA+ E D R GLQYGTNET +SD +
Sbjct: 715 PLNFDIYYDDPRSELLDGKLFSPVLAHIQPEFHYQANIESDGRYGLQYGTNETNMSDFLN 894
Query: 322 S-LTWDQLSYEESGSQPMNFPLFNVKDSGSGSDSDFEITNM 361
S + WDQ+ +E+ Q ++PLFNVKD+ SD D E+ NM
Sbjct: 895 SVVNWDQVPFEDPNCQQQSYPLFNVKDNILNSDLDSELANM 1017
Score = 225 bits (574), Expect = 4e-59
Identities = 130/231 (56%), Positives = 159/231 (68%)
Frame = +3
Query: 364 MQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSDVSM 423
MQ Y E AID +IPL EF S G+P D GDEQKSNVGLFQN + T DV+M
Sbjct: 1185 MQAGYAEGAIDGRIPLLITSEFCSTTGIPVDCVGDEQKSNVGLFQNNSQRAFT---DVNM 1355
Query: 424 GQVYNVVNDYEQPRNWNVATGGDTGIIIRARQAQNELVNTNINQGSANRRIRLLKSAHGS 483
GQVYNVV+DYEQ R + G+TGII R R+ +NE ++ N QG+A RRIRL + HGS
Sbjct: 1356 GQVYNVVDDYEQQRICSAVASGNTGIIRRHREVRNEQLSINSTQGTAQRRIRLSMATHGS 1535
Query: 484 SRVVKGGRRAQEDRNSKPITAVEKKASESLAADEDDTLTNHVKAMPKTPKSTNNRKIYQR 543
++ VK AQE+ +KP+ AV +K SE+ A+DE TLTN + KTP+ST RKI Q+
Sbjct: 1536 NKTVKSESCAQEELYAKPVIAVGEKGSENHASDESATLTNDGNELQKTPESTGKRKISQQ 1715
Query: 544 GSNAGSSTKVLKDLLLLRRVPCISKASSNRAMWSSVFVVSAFVMVSVMVFS 594
+ AG ST LKDL LLRRVP ISKASSNR SSVFV+SAFVMVS++VF+
Sbjct: 1716 VTKAG-STLGLKDLFLLRRVPHISKASSNRTKCSSVFVLSAFVMVSLVVFT 1865
>TC212360 similar to UP|O81790 (O81790) NAM / CUC2-like protein, partial
(26%)
Length = 575
Score = 288 bits (736), Expect = 6e-78
Identities = 133/159 (83%), Positives = 144/159 (89%), Gaps = 5/159 (3%)
Frame = +2
Query: 1 MGAVETAVP-----VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCK 55
MGAV P VLSLN+LP GFRFRP+DEE+ID+YLR KINGN D+VWVIREIDVCK
Sbjct: 98 MGAVVECYPPPRTAVLSLNTLPLGFRFRPTDEELIDYYLRSKINGNSDDVWVIREIDVCK 277
Query: 56 WEPWDLPDLSVIRNRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLI 115
WEPWDLPDLSV+RN+DPEWFFFCPQDRKYPNG+RLNRAT GYWKATGKDRKIKSGS+LI
Sbjct: 278 WEPWDLPDLSVVRNKDPEWFFFCPQDRKYPNGHRLNRATNHGYWKATGKDRKIKSGSTLI 457
Query: 116 GMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPG 154
GMKKTLVFY GRAPKGKRTNWVMHEYRPTL+EL+GTNPG
Sbjct: 458 GMKKTLVFYTGRAPKGKRTNWVMHEYRPTLKELEGTNPG 574
>TC206828 similar to UP|Q7VJ29 (Q7VJ29) Biotin operon repressor BirA, partial
(5%)
Length = 1428
Score = 218 bits (555), Expect = 6e-57
Identities = 159/365 (43%), Positives = 199/365 (53%), Gaps = 22/365 (6%)
Frame = +3
Query: 250 QPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDYQASNEPDDRSGLQY 309
Q Q V+ AEE+QPLN+D++ D N DDKLFSP H P F YQA+N+ S LQY
Sbjct: 3 QDQIVEPVAEEDQPLNVDIFYDPTNLPLDDKLFSPVHAHFPTDFYYQANNQ----SELQY 170
Query: 310 GTNETAISDLFDSLT-WDQLSYEESGSQPMNFPLFNVKDSGSGSDSDFEITNMPSMQI-- 366
GTNE IS+ FDS+ WD S++ S + L N D+GSGSDSD E+TNMP +Q
Sbjct: 171 GTNEN-ISEFFDSVVNWDAFSHDTSSLDLSSSFLINNTDNGSGSDSDVEMTNMPHLQALH 347
Query: 367 PYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSDVSMGQV 426
YP+EA+ EQKSNVGLFQN QM S+D +MGQV
Sbjct: 348 NYPKEAV-------------------------EQKSNVGLFQNN--FQMALSNDSTMGQV 446
Query: 427 YNVVNDYEQPRNWNVATGGDTGIIIRARQAQNELVNTNI---NQGSANRRIRL------L 477
+N+VN EQPRN++ DTGI IR R +NE N + +QG A RRIRL L
Sbjct: 447 HNMVNYCEQPRNFDAFVSADTGIRIRTRPGRNEQPNMDTMVQSQGIAPRRIRLGVVRRPL 626
Query: 478 KSAHGSSRVVKGGRRAQEDRNSKPITAVEKKASE---------SLAADEDDTLTNHV-KA 527
S GS A ED NSK I EKKASE S +D T+T+ V +
Sbjct: 627 GSQDGSCAC------APEDHNSKTIITAEKKASESENHAADGSSTVSDMSSTVTDDVDEP 788
Query: 528 MPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASSNRAMWSSVFVVSAFVM 587
+P+ST+ R I ++ S K LL+ RRV SK SSNRA WS+V VSA V+
Sbjct: 789 EEASPESTDLRNISEKHVATAESVAGSKVLLVNRRVHYTSKVSSNRATWSNVLAVSAAVL 968
Query: 588 VSVMV 592
VS ++
Sbjct: 969 VSAIL 983
>TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, complete
Length = 1216
Score = 180 bits (457), Expect = 1e-45
Identities = 106/253 (41%), Positives = 140/253 (54%), Gaps = 7/253 (2%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I YL +K+ I E+D+ + EPWDLP + + + EW+
Sbjct: 170 LPPGFRFHPTDEELISHYLYRKVTDTNFSARAIGEVDLNRSEPWDLPWKAKMGEK--EWY 343
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FFC +DRKYP G R NRAT GYWKATGKD++I G SL+GMKKTLVFY GRAPKG++T+
Sbjct: 344 FFCVRDRKYPTGLRTNRATESGYWKATGKDKEIFRGKSLVGMKKTLVFYKGRAPKGEKTD 523
Query: 136 WVMHEYRPTLQELDG-------TNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMA 188
WVMHEYR LDG +N +VICR+F+K ISG M
Sbjct: 524 WVMHEYR-----LDGKFSFHNLPKTAKNEWVICRVFQKSSSVKRTHISG-------MMML 667
Query: 189 AKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYDAQN 248
Y E V S + LP + SS + + K ++ SN P+D +D+ N
Sbjct: 668 DSYGNEMVYSSSALPPLTDSSPSIGNNTKALSVTDSAYVPCFSN---PIDVPRGIFDSLN 838
Query: 249 AQPQNVKLAAEEN 261
N+ ++ N
Sbjct: 839 ----NINISINSN 865
>TC206829 UP|Q93ZN0 (Q93ZN0) AT4g35580/F8D20_90, partial (5%)
Length = 811
Score = 178 bits (452), Expect = 5e-45
Identities = 128/291 (43%), Positives = 158/291 (53%), Gaps = 18/291 (6%)
Frame = +2
Query: 271 DLKNDIFDDKLFSPALVHLPPVFDYQASNEPDDRSGLQYGTNETAISDLFDSLT-WDQLS 329
D N DDKLFSP H P F YQA+N+ S LQYGT+E IS+ FDS+ WD S
Sbjct: 2 DPTNQQLDDKLFSPVHAHFPADFYYQANNQ----SELQYGTSEN-ISEFFDSVVNWDSFS 166
Query: 330 YEESGSQPMNFPLFNVKDSGSGSDSDFEITNMPSMQI--PYPEEAIDMKIPLGTAPEFFS 387
Y S S + N KD+GSGSDSD E+ NM +Q YP+EA+
Sbjct: 167 YATS-SLXLGSSFLNPKDNGSGSDSDVEMANMLHLQALHDYPKEAV-------------- 301
Query: 388 PVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSDVSMGQVYNVVNDYEQPRNWNVATGGDT 447
E+K NVGLFQN QM FS+D MGQV+NV N EQPRN++ GDT
Sbjct: 302 -----------EEKFNVGLFQNN--PQMAFSNDGPMGQVHNVANYCEQPRNFDAFVNGDT 442
Query: 448 GIIIRARQAQNELVNTNI---NQGSANRRIRL--LKSAHGSSRVVKGG--RRAQEDRNSK 500
GI IR R +NE N N+ +QG A RRIRL ++ GS K G A ED NSK
Sbjct: 443 GIRIRTRHGRNEQPNMNMMIQSQGIAPRRIRLGVVRRPLGSQETAKDGSCACAPEDHNSK 622
Query: 501 PITAVEKKASESLAADEDDTLTNHVKAM--------PKTPKSTNNRKIYQR 543
I + EKKASE+ AADE T+++ + +PKST+ RKI Q+
Sbjct: 623 TIISAEKKASENHAADESSTVSDESSTVTDDSDEPEEASPKSTDPRKISQQ 775
>TC232291 similar to UP|O81790 (O81790) NAM / CUC2-like protein, partial
(20%)
Length = 716
Score = 174 bits (441), Expect = 1e-43
Identities = 83/162 (51%), Positives = 108/162 (66%), Gaps = 2/162 (1%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
+P GFRFRP+DEE++D+YL+ K+ + V +I EID+CK EPWD+P SVI++ DPEWF
Sbjct: 181 MPVGFRFRPTDEELVDYYLKHKLLADDFPVHIIPEIDLCKVEPWDVPGRSVIKSDDPEWF 360
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDR--KIKSGSSLIGMKKTLVFYHGRAPKGKR 133
FF P D KY R NR T GYWK TG DR KI S++IG KKTLVF+ GR P+G +
Sbjct: 361 FFSPVDYKYLKSKRFNRTTKRGYWKTTGNDRNVKIPGTSNVIGTKKTLVFHEGRGPRGVK 540
Query: 134 TNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSI 175
TNWV+HEY ++ Q +V+CRL KK ++ E I
Sbjct: 541 TNWVIHEYHAVT-----SHESQRAFVLCRLMKKAEKKNEGGI 651
>TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein,
partial (58%)
Length = 935
Score = 171 bits (433), Expect = 8e-43
Identities = 88/166 (53%), Positives = 112/166 (67%), Gaps = 4/166 (2%)
Frame = +3
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I YL +K+ I E+D+ K EPWDLP + + + EW+
Sbjct: 192 LPPGFRFHPTDEELISHYLYKKVIDTKFCARAIGEVDLNKSEPWDLPWKAKMGEK--EWY 365
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FFC +DRKYP G R NRAT GYWKATGKD++I G SL+GMKKTLVFY GRAPKG+++N
Sbjct: 366 FFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFRGKSLVGMKKTLVFYRGRAPKGEKSN 545
Query: 136 WVMHEYRP----TLQELDGTNPGQNPYVICRLFKKQDESIEVSISG 177
WVMHEYR ++ L T +N +VICR+F+K + ISG
Sbjct: 546 WVMHEYRLEGKFSVHNLPKT--AKNEWVICRVFQKSSAGKKTHISG 677
>TC215244 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (57%)
Length = 1463
Score = 160 bits (406), Expect = 1e-39
Identities = 77/167 (46%), Positives = 110/167 (65%)
Frame = +1
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
LS SLP GFRF P+DEE++ YL +K+ G+ + +I EID+ K++PW LP ++ +
Sbjct: 103 LSQLSLPPGFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEIDLYKFDPWVLPSKAIFGEK 282
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
EW+FF P+DRKYPNG+R NR GYWKATG D+ I + +G+KK LVFY G+APK
Sbjct: 283 --EWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYVGKAPK 456
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISG 177
G +TNW+MHEYR T + +V+CR++KK + + + +G
Sbjct: 457 GTKTNWIMHEYRLLDSSRKNTGTKLDDWVLCRIYKKNSSAQKTAQNG 597
>TC215247 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (46%)
Length = 643
Score = 159 bits (403), Expect = 3e-39
Identities = 76/157 (48%), Positives = 106/157 (67%)
Frame = +2
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
LS SLP GFRF P+DEE++ YL +K+ G+ + +I EID+ K++PW LP ++ +
Sbjct: 137 LSQLSLPPGFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEIDLYKFDPWVLPSKAIFGEK 316
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
EW+FF P+DRKYPNG+R NR GYWKATG D+ I + +G+KK LVFY G+APK
Sbjct: 317 --EWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYIGKAPK 490
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQ 167
G +TNW+MHEYR T + +V+CR++KK+
Sbjct: 491 GTKTNWIMHEYRLLDSSRKNTGTKLDDWVLCRIYKKK 601
>TC205765
Length = 749
Score = 159 bits (402), Expect = 3e-39
Identities = 88/156 (56%), Positives = 114/156 (72%)
Frame = +3
Query: 439 WNVATGGDTGIIIRARQAQNELVNTNINQGSANRRIRLLKSAHGSSRVVKGGRRAQEDRN 498
++ G+TGII R R+ QNE +++N QG+A RRIRL + HGS+++VK G AQE+ N
Sbjct: 18 YSALPSGNTGIITRHREVQNEQLSSNSTQGTAQRRIRLSMATHGSNKMVKSGSCAQEELN 197
Query: 499 SKPITAVEKKASESLAADEDDTLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLL 558
K + A+E+KASES +DE TLTN +PKT KS+ RKI ++ + AG ST LKD L
Sbjct: 198 LKSVIAMEEKASESHISDESSTLTNDGNELPKTSKSSGKRKISRQATKAG-STLGLKDFL 374
Query: 559 LLRRVPCISKASSNRAMWSSVFVVSAFVMVSVMVFS 594
LLRRVP ISKASSNRA SSVFVVSAFVMV+++VF+
Sbjct: 375 LLRRVPYISKASSNRAKCSSVFVVSAFVMVTLVVFT 482
>TC215245 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (58%)
Length = 1597
Score = 155 bits (393), Expect = 4e-38
Identities = 79/189 (41%), Positives = 119/189 (62%), Gaps = 5/189 (2%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE++ YL +K+ G+ + +I E+D+ K++PW LP + + EW
Sbjct: 152 SLPPGFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEVDLYKFDPWVLPGKAAFGEK--EW 325
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P+DRKYPNG+R NR GYWKATG D+ I + +G+KK LVFY G+APKG +T
Sbjct: 326 YFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYIGKAPKGSKT 505
Query: 135 NWVMHEYRPTLQELDGTNPGQ---NPYVICRLFKKQD--ESIEVSISGEAEQTASTPMAA 189
NW+MHEYR L N G + +V+CR++KK + +E ++ S+P ++
Sbjct: 506 NWIMHEYR-LLDSSRKHNLGTAKLDDWVLCRIYKKNSSAQKVEANLLAMECSNGSSPSSS 682
Query: 190 KYSPEEVQS 198
+ + ++S
Sbjct: 683 SHVDDMLES 709
>TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, partial
(89%)
Length = 1237
Score = 155 bits (392), Expect = 5e-38
Identities = 80/182 (43%), Positives = 113/182 (61%), Gaps = 11/182 (6%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I +YL + +I E+D+ K++PW+LP+ + + EW+
Sbjct: 182 LPPGFRFHPTDEELIVYYLCNQATSRPCPASIIPEVDIYKFDPWELPEKTDFGEK--EWY 355
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P++RKYPNG R NRAT+ GYWKATG D+ I SGS +G+KK LVFY G+ PKG +T+
Sbjct: 356 FFSPRERKYPNGVRPNRATVSGYWKATGTDKAIYSGSKHVGVKKALVFYKGKPPKGLKTD 535
Query: 136 WVMHEYRPTLQELDGTNPGQN---------PYVICRLFKKQD--ESIEVSISGEAEQTAS 184
W+MHEYR L G+ N +V+CR++KK++ +S+E Q
Sbjct: 536 WIMHEYR-----LIGSRRQANRQVGSMRLDDWVLCRIYKKKNIGKSMEAKEEYPIAQINL 700
Query: 185 TP 186
TP
Sbjct: 701 TP 706
>TC211321 homologue to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, partial
(57%)
Length = 612
Score = 155 bits (391), Expect = 6e-38
Identities = 70/129 (54%), Positives = 92/129 (71%)
Frame = +1
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPE 73
+ LP GFRF P+DEE+I +YL + +I E+D+ K++PW+LPD + E
Sbjct: 151 SELPPGFRFHPTDEELIVYYLCNQATSKPCPASIIPEVDLYKFDPWELPDKTEFGEN--E 324
Query: 74 WFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKR 133
W+FF P+DRKYPNG R NRAT+ GYWKATG D+ I SGS +G+KK+LVFY GR PKG +
Sbjct: 325 WYFFSPRDRKYPNGVRPNRATVSGYWKATGTDKAIYSGSKNVGVKKSLVFYKGRPPKGAK 504
Query: 134 TNWVMHEYR 142
T+W+MHEYR
Sbjct: 505 TDWIMHEYR 531
>TC226098 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (45%)
Length = 1562
Score = 155 bits (391), Expect = 6e-38
Identities = 106/284 (37%), Positives = 150/284 (52%), Gaps = 36/284 (12%)
Frame = +1
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPE 73
++LP GFRF P+DEE+I YL +K+ V +I E+D+ K +PWDLP + + E
Sbjct: 163 SNLPPGFRFHPTDEELILHYLSKKVASIPLPVSIIAEVDIYKLDPWDLPAKATFGEK--E 336
Query: 74 WFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKS-----GSSLIGMKKTLVFYHGRA 128
W+FF P+DRKYPNG R NRA GYWKATG D+ I + +G+KK LVFY GR
Sbjct: 337 WYFFSPRDRKYPNGARPNRAAASGYWKATGTDKTIVTSLQGGAQESVGVKKALVFYKGRP 516
Query: 129 PKGKRTNWVMHEYRPTLQ----ELDGTNPGQNPYVICRLFKKQDE---SIEVSIS-GEAE 180
PKG +TNW+MHEYR +L ++ + +V+CR++KK S E SI GE +
Sbjct: 517 PKGVKTNWIMHEYRLVDNNKPIKLKDSSMRLDDWVLCRIYKKSKHPLTSTEASIGVGEVD 696
Query: 181 QT-----ASTPMAAKYSPEEVQSDTPLPLVAVS-------------SSLLTED-DKHQAI 221
Q T + SP +T + +VS SS L+E+ H
Sbjct: 697 QAEEHVFKETLLPISKSPTPPPQNTLISQKSVSFSNLLDAMDYSILSSFLSENHTTHPPG 876
Query: 222 IPETSEETTSNI---ITPVDRNSDRYDAQNAQPQ-NVKLAAEEN 261
+ ++ T N+ +P+D NS+ Y Q PQ N ++ EN
Sbjct: 877 VGSSTGFNTGNLDQQPSPID-NSNGYMFQKNNPQLNPSVSNMEN 1005
>TC226101 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (66%)
Length = 825
Score = 154 bits (390), Expect = 8e-38
Identities = 83/191 (43%), Positives = 120/191 (62%), Gaps = 9/191 (4%)
Frame = +3
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+D+E+++ YL +K V +I+EID+ K++PW LP++++ + EW+
Sbjct: 54 LPPGFRFHPTDDELVNHYLCRKCAAQTIAVPIIKEIDLYKFDPWQLPEMALYGEK--EWY 227
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG+R NRA GYWKATG D+ I +L G+KK LVFY G+APKG +TN
Sbjct: 228 FFSPRDRKYPNGSRPNRAAGSGYWKATGADKPIGKPKAL-GIKKALVFYAGKAPKGVKTN 404
Query: 136 WVMHEYRPTLQELDGTNPGQN---------PYVICRLFKKQDESIEVSISGEAEQTASTP 186
W+MHEYR L +D + +N +V+CR++ K+ + + + +Q
Sbjct: 405 WIMHEYR--LANVDRSASKKNTTTNNLRLDDWVLCRIYNKKGKIEKYNGVAVVDQ----- 563
Query: 187 MAAKYSPEEVQ 197
AK S EEVQ
Sbjct: 564 KVAKLSEEEVQ 596
>TC227622 similar to UP|Q7XJT9 (Q7XJT9) NAM (No apical meristem)-like
protein, partial (61%)
Length = 1113
Score = 154 bits (389), Expect = 1e-37
Identities = 98/289 (33%), Positives = 150/289 (50%), Gaps = 16/289 (5%)
Frame = +2
Query: 19 GFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWFFFC 78
GFRF P++EE++DFYL+ + G VI +++ + +PWDLP L+ + R EW+FF
Sbjct: 137 GFRFHPTEEELLDFYLKNMVVGKKLRYDVIGFLNIYQHDPWDLPGLAKVGER--EWYFFV 310
Query: 79 PQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSS---LIGMKKTLVFYHGRAPKGKRTN 135
P+D+K+ +G R NR T G+WKATG DRKI + S +IG++KTLVFY GRAP+G +T+
Sbjct: 311 PRDKKHGSGGRPNRTTEKGFWKATGSDRKIVTLSDPKRIIGLRKTLVFYQGRAPRGCKTD 490
Query: 136 WVMHEYRPTLQELDGTNPGQNPYVICRLFKK-------------QDESIEVSISGEAEQT 182
WVM+EYR L V+C++++K ++E + G
Sbjct: 491 WVMNEYR-----LPDNCKLPKEIVLCKIYRKATSLKVLEQRAALEEEREMKQMVGSPASP 655
Query: 183 ASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSD 242
+ST + SP+E Q+ +PL + L E + ++P+ +E +N + D N
Sbjct: 656 SSTDTMSFSSPQEEQN---MPLPLLQHVLKKESETEDMVVPK--QEKITNQLVLKDTNKS 820
Query: 243 RYDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPP 291
P E+ L L M +D D F +L SP L + P
Sbjct: 821 TCGTSLQMP----FGKEKLPDLQLPMLSDWTQDTFWAQLNSPWLQNFTP 955
>TC226284 similar to UP|Q9FY93 (Q9FY93) NAM-like protein
(AT5g13180/T19L5_140), partial (59%)
Length = 1601
Score = 152 bits (385), Expect = 3e-37
Identities = 73/154 (47%), Positives = 102/154 (65%), Gaps = 2/154 (1%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE++ YL++K+ +I E+DVCK +PWDLP + + E +
Sbjct: 215 LPPGFRFHPTDEELVLQYLKRKVFSCPLPASIIPEVDVCKSDPWDLPG-----DLEQERY 379
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKI--KSGSSLIGMKKTLVFYHGRAPKGKR 133
FF ++ KYPNGNR NRAT GYWKATG D++I G+ ++GMKKTLVFY G+ P G R
Sbjct: 380 FFSTKEAKYPNGNRSNRATNSGYWKATGLDKQIVTSKGNQVVGMKKTLVFYRGKPPHGSR 559
Query: 134 TNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQ 167
T+W+MHEYR + ++ +V+CR+F K+
Sbjct: 560 TDWIMHEYRLNILNASQSHVPMENWVLCRIFLKK 661
>TC216430 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1, complete
Length = 1289
Score = 152 bits (384), Expect = 4e-37
Identities = 79/181 (43%), Positives = 113/181 (61%), Gaps = 7/181 (3%)
Frame = +3
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P DEE++ YL +K+ N + + +D+ K EPWD+P+ + + + EW+
Sbjct: 141 LPPGFRFHPRDEELVCDYLMKKVAHNDSLLMI--NVDLNKCEPWDIPETACVGGK--EWY 308
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
F+ +DRKY G R NRAT GYWKATGKDR I +L+GM+KTLVFY GRAPKGK+T
Sbjct: 309 FYTQRDRKYATGLRTNRATASGYWKATGKDRSILRKGTLVGMRKTLVFYQGRAPKGKKTE 488
Query: 136 WVMHEYRPTLQELDG------TNPGQNPYVICRLF-KKQDESIEVSISGEAEQTASTPMA 188
WVMHE+R ++G + + +V+CR+F K ++ S + + E T S+ +
Sbjct: 489 WVMHEFR-----IEGPHGPPKISSSKEDWVLCRVFYKNREVSAKPRMGSCYEDTGSSSLP 653
Query: 189 A 189
A
Sbjct: 654 A 656
>TC215363 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (64%)
Length = 1464
Score = 151 bits (381), Expect = 9e-37
Identities = 76/157 (48%), Positives = 105/157 (66%), Gaps = 5/157 (3%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE++ YL +K V +I EID+ K++PWDLP L+ + EW+
Sbjct: 79 LPPGFRFHPTDEELVLHYLCRKCASQPIAVPIIAEIDLYKYDPWDLPGLATYGEK--EWY 252
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG+R NRA GYWKATG D+ I +G+KK LVFY G+APKG ++N
Sbjct: 253 FFSPRDRKYPNGSRPNRAAGTGYWKATGADKPIGQPKP-VGIKKALVFYAGKAPKGDKSN 429
Query: 136 WVMHEYRPTLQELDGTNPGQN-----PYVICRLFKKQ 167
W+MHEYR L ++D + +N +V+CR++ K+
Sbjct: 430 WIMHEYR--LADVDRSVRKKNTLRLDDWVLCRIYNKK 534
>TC215440 homologue to UP|Q7Y1A6 (Q7Y1A6) NAC-domain protein 18, partial
(70%)
Length = 983
Score = 151 bits (381), Expect = 9e-37
Identities = 83/225 (36%), Positives = 127/225 (55%), Gaps = 5/225 (2%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE++ YL +K V +I EID+ K++PWDLP +++ + EW+
Sbjct: 91 LPPGFRFHPTDEELVVHYLCRKCASQEIAVPIIAEIDLYKYDPWDLPGMALYGEK--EWY 264
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG+R NR+ GYWKATG D+ + +G+KK LVFY G+APKG +TN
Sbjct: 265 FFTPRDRKYPNGSRPNRSAGTGYWKATGADKPVGKPKP-VGIKKALVFYAGKAPKGVKTN 441
Query: 136 WVMHEYRPTLQELDGTNPGQN-----PYVICRLFKKQDESIEVSISGEAEQTASTPMAAK 190
W+MHEYR L ++D + +N +V+CR++ K+ + E +Q A P +
Sbjct: 442 WIMHEYR--LADVDRSVRKKNSLRLDDWVLCRIYNKKG-------AIEKQQPAPPPPSGV 594
Query: 191 YSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIIT 235
+ E + + P + L E + T + +++
Sbjct: 595 HKIECYEMEDVKPEYTAADCLYFEASDSVPRLHTTESSCSEQVVS 729
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,570,788
Number of Sequences: 63676
Number of extensions: 311343
Number of successful extensions: 1294
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 1193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1197
length of query: 596
length of database: 12,639,632
effective HSP length: 103
effective length of query: 493
effective length of database: 6,081,004
effective search space: 2997934972
effective search space used: 2997934972
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0207.16


