

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
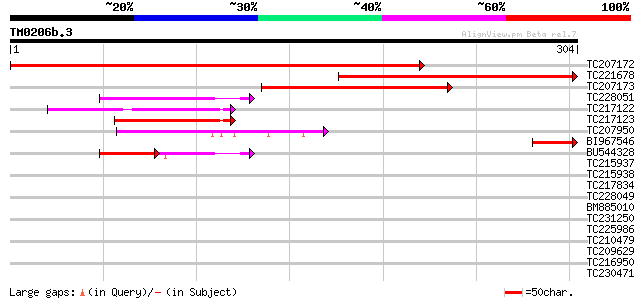
Query= TM0206b.3
(304 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207172 417 e-117
TC221678 198 3e-51
TC207173 189 1e-48
TC228051 similar to UP|Q91YE7 (Q91YE7) RNA binding motif protein... 54 1e-07
TC217122 similar to UP|Q94C11 (Q94C11) AT3g52120/F4F15_230, part... 51 7e-07
TC217123 similar to UP|Q94C11 (Q94C11) AT3g52120/F4F15_230, part... 50 2e-06
TC207950 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {A... 44 7e-05
BI967546 43 2e-04
BU544328 35 2e-04
TC215937 similar to UP|Q940M0 (Q940M0) At1g63980/F22C12_9, parti... 40 0.001
TC215938 similar to UP|Q940M0 (Q940M0) At1g63980/F22C12_9, parti... 40 0.001
TC217834 similar to UP|D111_ARATH (P42698) DNA-damage-repair/tol... 40 0.002
TC228049 37 0.011
BM885010 similar to GP|5702039|emb| Fuct c3 protein {Vigna radia... 36 0.019
TC231250 similar to UP|Q9ST51 (Q9ST51) Fuct c3 protein, partial ... 35 0.055
TC225986 weakly similar to UP|Q9W1I6 (Q9W1I6) CG18426-PA (LD1203... 34 0.072
TC210479 similar to UP|Q94EI2 (Q94EI2) AT5g08540/MAH20_10, parti... 33 0.16
TC209629 similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein (AT3g098... 33 0.21
TC216950 33 0.21
TC230471 similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein (AT3g098... 32 0.27
>TC207172
Length = 809
Score = 417 bits (1071), Expect = e-117
Identities = 203/222 (91%), Positives = 218/222 (97%)
Frame = +2
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASL 60
M+YRR+SGRQE+GGR RRQ K+E+A+QDSLI DL++DFRLPINHRPTENVDL+NVEQASL
Sbjct: 143 MEYRRTSGRQETGGRDRRQSKREQAYQDSLIEDLSQDFRLPINHRPTENVDLDNVEQASL 322
Query: 61 DTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEEN 120
DTQ+TSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSG+RDPRLG+GKQEEDDFFTAEEN
Sbjct: 323 DTQITSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGIRDPRLGVGKQEEDDFFTAEEN 502
Query: 121 IQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 180
IQRKKLDVELEETEE+V+KREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL
Sbjct: 503 IQRKKLDVELEETEEHVKKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 682
Query: 181 SSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFA 222
SSYDHNHRKRFKQMKEMHG SSRDDRQKREQQRQERE+AKFA
Sbjct: 683 SSYDHNHRKRFKQMKEMHGSSSRDDRQKREQQRQEREMAKFA 808
>TC221678
Length = 791
Score = 198 bits (503), Expect = 3e-51
Identities = 101/128 (78%), Positives = 108/128 (83%)
Frame = +2
Query: 177 EAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLL 236
EAHL YD NHRK FKQ EMHGGSSRDDRQKREQ RQERE+AKFAQIADAQKQQ+LQL
Sbjct: 5 EAHLXXYDXNHRKXFKQXXEMHGGSSRDDRQKREQLRQEREMAKFAQIADAQKQQQLQLQ 184
Query: 237 QESGSEPVSSETRTATPLTDQEQRNALKFGFSSKGSASKNVIGPKRQMVAKKQNIPVSSI 296
QE GS V SE+ TAT LTDQEQRN LKFGFSSKGSASK G K+Q VAKKQN+P+SSI
Sbjct: 185 QEXGSATVPSESITATALTDQEQRNTLKFGFSSKGSASKITFGAKKQNVAKKQNVPISSI 364
Query: 297 FNNDSDEE 304
F+NDSDEE
Sbjct: 365 FSNDSDEE 388
>TC207173
Length = 595
Score = 189 bits (481), Expect = 1e-48
Identities = 93/102 (91%), Positives = 98/102 (95%)
Frame = +3
Query: 136 NVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK 195
++ K +VLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK
Sbjct: 288 HIMKIQVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK 467
Query: 196 EMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQ 237
EMHGGSSRDDRQKREQ RQERE+AKFAQIADAQKQQ+LQL Q
Sbjct: 468 EMHGGSSRDDRQKREQLRQEREMAKFAQIADAQKQQQLQLQQ 593
Score = 35.0 bits (79), Expect = 0.042
Identities = 16/19 (84%), Positives = 18/19 (94%)
Frame = +1
Query: 124 KKLDVELEETEENVRKREV 142
K LDVELEETEE+V+KREV
Sbjct: 4 KNLDVELEETEEHVKKREV 60
>TC228051 similar to UP|Q91YE7 (Q91YE7) RNA binding motif protein 5, partial
(3%)
Length = 1309
Score = 53.5 bits (127), Expect = 1e-07
Identities = 30/84 (35%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Frame = +1
Query: 49 NVDLENVEQASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLG 107
+ +L+ E + D + +N+G ++L+ MGW+ G GLGKD G+IEP+ + + R GLG
Sbjct: 865 DANLDTFEVITADKAIDENNVGNRMLRNMGWQEGLGLGKDGSGMIEPVLAQATENRAGLG 1044
Query: 108 KQEEDDFFTAEENIQRKKLDVELE 131
Q+ KKLD LE
Sbjct: 1045SQQ-------------KKLDPSLE 1077
>TC217122 similar to UP|Q94C11 (Q94C11) AT3g52120/F4F15_230, partial (38%)
Length = 914
Score = 50.8 bits (120), Expect = 7e-07
Identities = 35/103 (33%), Positives = 54/103 (51%), Gaps = 2/103 (1%)
Frame = +3
Query: 21 KKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASLDTQLTSSNIGFKLLQKMGWK 80
KK D + + E F N + E E+A + + N+G +LL KMGWK
Sbjct: 141 KKGHHMGDFIPPEELEKFLATCNDAAAQKAAREAAERAKIQ----ADNVGHRLLSKMGWK 308
Query: 81 -GKGLGKDEQGIIEPIKSG-MRDPRLGLGKQEEDDFFTAEENI 121
G+GLG +GI +PI +G ++ LG+G QE + +AE++I
Sbjct: 309 EGEGLGGSRKGIADPIMAGNVKKNNLGVGAQEPGE-VSAEDDI 434
>TC217123 similar to UP|Q94C11 (Q94C11) AT3g52120/F4F15_230, partial (23%)
Length = 678
Score = 49.7 bits (117), Expect = 2e-06
Identities = 27/67 (40%), Positives = 45/67 (66%), Gaps = 2/67 (2%)
Frame = +2
Query: 57 QASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSG-MRDPRLGLGKQEEDDF 114
+A+ ++ + N+G +LL KMGWK G+GLG +GI +PI +G ++ LG+G QE +
Sbjct: 23 EAAERAKIQADNVGHRLLSKMGWKEGEGLGGSRKGIADPIMAGNVKKNNLGVGAQEPGE- 199
Query: 115 FTAEENI 121
+AE++I
Sbjct: 200 VSAEDDI 220
>TC207950 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {Arabidopsis
thaliana;} , partial (72%)
Length = 1259
Score = 44.3 bits (103), Expect = 7e-05
Identities = 44/160 (27%), Positives = 71/160 (43%), Gaps = 46/160 (28%)
Frame = +1
Query: 58 ASLDTQLTSSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLGLG--------- 107
A ++ + SNIGFKLL++MG+ G LGK G EP+ +R R G+G
Sbjct: 253 AKVEAPIPQSNIGFKLLKQMGYTPGSALGKQGSGRAEPVGIEIRRSRAGVGLEDPHKEKR 432
Query: 108 -----------KQEED---DFFTAEE----------NIQRKKLDVELEETEENV--RKRE 141
++EED +F + ++ N + K ++ E E V +K +
Sbjct: 433 KNEGIMMDRKRRKEEDLMKEFGSRQKSQWQSRRVIINYNKAKAALDQLENREIVEPQKND 612
Query: 142 VLAEREQKIQTEVKE---------IRKVF-YCDLCNKQYK 171
+AE E++ + E+ E +R F YC C QY+
Sbjct: 613 DVAEGEEEEEEEITEEDLHDVLMKLRDEFNYCLFCGCQYE 732
>BI967546
Length = 309
Score = 43.1 bits (100), Expect = 2e-04
Identities = 18/24 (75%), Positives = 23/24 (95%)
Frame = -1
Query: 281 KRQMVAKKQNIPVSSIFNNDSDEE 304
K+Q VAKKQN+P+SSIF+NDSDE+
Sbjct: 306 KKQNVAKKQNVPISSIFSNDSDED 235
>BU544328
Length = 474
Score = 34.7 bits (78), Expect(2) = 2e-04
Identities = 23/61 (37%), Positives = 28/61 (45%), Gaps = 8/61 (13%)
Frame = -1
Query: 79 WKGK--------GLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVEL 130
W GK GLGKD G+IEP+ + + R GLG Q+ KKLD L
Sbjct: 339 WAGKKD**FDAQGLGKDGSGMIEPVLAQATENRAGLGSQQ-------------KKLDPSL 199
Query: 131 E 131
E
Sbjct: 198 E 196
Score = 27.3 bits (59), Expect(2) = 2e-04
Identities = 9/32 (28%), Positives = 20/32 (62%)
Frame = -3
Query: 49 NVDLENVEQASLDTQLTSSNIGFKLLQKMGWK 80
+ +L+ E + D + +N+G ++L+ MGW+
Sbjct: 424 DANLDTFEVITADKAIDENNVGNRMLRNMGWQ 329
>TC215937 similar to UP|Q940M0 (Q940M0) At1g63980/F22C12_9, partial (28%)
Length = 460
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/77 (29%), Positives = 47/77 (60%), Gaps = 3/77 (3%)
Frame = +2
Query: 71 FKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD--FFTAEENIQRKKLD 127
F+L+++MGW+ G+GLGK++QGI ++ + +G+G ++ ++ F T + + K+L
Sbjct: 116 FRLMKQMGWEEGEGLGKEKQGIKGHVRVKNKQDTIGIGLEKPNNWAFDTTQFDNILKRLK 295
Query: 128 VELEETEENVRKREVLA 144
V+ ++ + K E A
Sbjct: 296 VQAPQSHDIEEKVETKA 346
>TC215938 similar to UP|Q940M0 (Q940M0) At1g63980/F22C12_9, partial (60%)
Length = 1505
Score = 40.0 bits (92), Expect = 0.001
Identities = 23/86 (26%), Positives = 52/86 (59%), Gaps = 3/86 (3%)
Frame = +3
Query: 71 FKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD--FFTAEENIQRKKLD 127
F+L+++MGW+ G+GLGK++QGI ++ + +G+G ++ ++ F T + + K+L
Sbjct: 108 FRLMKQMGWEEGEGLGKEKQGIKGHVRVKSKHDTIGIGLEKPNNWAFDTTQFDNILKRLK 287
Query: 128 VELEETEENVRKREVLAEREQKIQTE 153
V+ ++ + + E+K++T+
Sbjct: 288 VQAPQSHD--------TDIEEKVETK 341
>TC217834 similar to UP|D111_ARATH (P42698) DNA-damage-repair/toleration
protein DRT111, chloroplast precursor, partial (57%)
Length = 1006
Score = 39.7 bits (91), Expect = 0.002
Identities = 18/36 (50%), Positives = 25/36 (69%), Gaps = 1/36 (2%)
Frame = +2
Query: 72 KLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGL 106
+++ KMGWK G+GLGK EQGI P+ + D R G+
Sbjct: 227 RMMAKMGWKEGQGLGKQEQGITTPLMAKKTDRRAGV 334
>TC228049
Length = 547
Score = 37.0 bits (84), Expect = 0.011
Identities = 21/52 (40%), Positives = 28/52 (53%)
Frame = +2
Query: 80 KGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELE 131
+G+GLGKD G+IEP+ + + R GLG Q+ KKLD LE
Sbjct: 185 EGQGLGKDGSGMIEPVLAQATENRAGLGSQQ-------------KKLDPSLE 301
>BM885010 similar to GP|5702039|emb| Fuct c3 protein {Vigna radiata var.
radiata}, partial (7%)
Length = 422
Score = 36.2 bits (82), Expect = 0.019
Identities = 18/25 (72%), Positives = 20/25 (80%), Gaps = 1/25 (4%)
Frame = +2
Query: 66 SSNIGFKLLQKMGWK-GKGLGKDEQ 89
SSNIGF+LL+K GWK G GLG EQ
Sbjct: 200 SSNIGFQLLKKHGWKEGTGLGVSEQ 274
>TC231250 similar to UP|Q9ST51 (Q9ST51) Fuct c3 protein, partial (7%)
Length = 399
Score = 34.7 bits (78), Expect = 0.055
Identities = 17/24 (70%), Positives = 19/24 (78%), Gaps = 1/24 (4%)
Frame = +2
Query: 67 SNIGFKLLQKMGWK-GKGLGKDEQ 89
SNIGF+LL+K GWK G GLG EQ
Sbjct: 8 SNIGFQLLKKHGWKEGTGLGVSEQ 79
>TC225986 weakly similar to UP|Q9W1I6 (Q9W1I6) CG18426-PA (LD12035p), partial
(13%)
Length = 1553
Score = 34.3 bits (77), Expect = 0.072
Identities = 44/186 (23%), Positives = 76/186 (40%), Gaps = 7/186 (3%)
Frame = +2
Query: 118 EENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIR-KVFYCDLCNKQYKLAMEF 176
+ ++R+K D ELE E + V EREQ+ + E E + + + D ++ + E
Sbjct: 29 DRELKREK-DRELERYEREAERERVRKEREQRRRVEEAERQYETYLKDWEYREREKEKER 205
Query: 177 EAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQK-----REQQRQEREIAKFAQIADAQKQQ 231
+ RKR K++ D R++ E++R++R K + D QK+
Sbjct: 206 QYEKEKEKERERKRRKEILYDEEDEDEDSRKRWRRSVIEEKRKKRLREKEDDLVDRQKE- 382
Query: 232 RLQLLQESGSEPVSSETRTATPLTDQEQRNALKFGFSSKGSASKNVI-GPKRQMVAKKQN 290
E +E + + QR+ALK S++V+ G K M+
Sbjct: 383 ----------EEEIAEAKKKAEEEQKRQRDALKL-------LSEHVVNGGKENMITDDIT 511
Query: 291 IPVSSI 296
V SI
Sbjct: 512 NEVKSI 529
>TC210479 similar to UP|Q94EI2 (Q94EI2) AT5g08540/MAH20_10, partial (33%)
Length = 671
Score = 33.1 bits (74), Expect = 0.16
Identities = 19/77 (24%), Positives = 40/77 (51%)
Frame = +2
Query: 83 GLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREV 142
GLG EQG +EP+++ +++ + GLG + + Q ++ +K +
Sbjct: 2 GLGVSEQGRLEPVETHVKNNKRGLGADKAKKKVVKAKPDQSDSSKGNNQQDHLPQKKSKT 181
Query: 143 LAEREQKIQTEVKEIRK 159
L++R +K+Q K++R+
Sbjct: 182 LSKRMRKMQEFEKKMRE 232
>TC209629 similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein
(AT3g09850/F8A24_10), partial (9%)
Length = 668
Score = 32.7 bits (73), Expect = 0.21
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = +1
Query: 66 SSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLGLG 107
+ G K++ KMG+ +G GLG++ QGI P+ + GLG
Sbjct: 202 TKGFGSKMMAKMGFVEGNGLGRESQGITTPLSAVRLPKSRGLG 330
>TC216950
Length = 873
Score = 32.7 bits (73), Expect = 0.21
Identities = 29/120 (24%), Positives = 54/120 (44%), Gaps = 21/120 (17%)
Frame = +2
Query: 118 EENIQRKKLDVELEETEENVRKR---EVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAM 174
EE+++ +++ VE+E E RKR EV A+ E++ + + E ++ K+ +
Sbjct: 281 EESLKSEEVQVEIERRLEEGRKRLNDEVAAQLEKEKEAALIEAKQ--------KEEQARK 436
Query: 175 EFEAHLSSYDHNHRK------------------RFKQMKEMHGGSSRDDRQKREQQRQER 216
E E + N RK R+K+++EM R+K+ ++ QER
Sbjct: 437 EKEDLERMLEENRRKVEEAQRREALEQQRREEERYKELEEMQRQKEEAMRKKKHEEEQER 616
>TC230471 similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein
(AT3g09850/F8A24_10), partial (10%)
Length = 660
Score = 32.3 bits (72), Expect = 0.27
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = +1
Query: 66 SSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLGLG 107
+ G K++ KMG+ +G GLG++ QGI P+ + GLG
Sbjct: 247 TKGFGSKMMAKMGFVEGTGLGRESQGITTPLSAVRLPKSRGLG 375
Score = 30.4 bits (67), Expect = 1.0
Identities = 14/28 (50%), Positives = 19/28 (67%)
Frame = +1
Query: 80 KGKGLGKDEQGIIEPIKSGMRDPRLGLG 107
+G GLGK+ QG+ +PI+ R LGLG
Sbjct: 13 EGAGLGKNGQGMAQPIEVIQRPKSLGLG 96
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.130 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,368,091
Number of Sequences: 63676
Number of extensions: 101403
Number of successful extensions: 718
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 714
length of query: 304
length of database: 12,639,632
effective HSP length: 97
effective length of query: 207
effective length of database: 6,463,060
effective search space: 1337853420
effective search space used: 1337853420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0206b.3


