

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
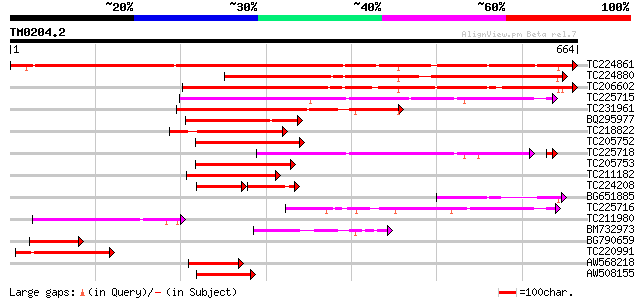
Query= TM0204.2
(664 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224861 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription... 1023 0.0
TC224880 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription... 598 e-171
TC206602 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription... 510 e-145
TC225715 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6... 357 1e-98
TC231961 homologue to GB|AAM16218.1|20334714|AY093957 At2g41900/... 322 3e-88
BQ295977 similar to GP|8843784|dbj zinc finger transcription fac... 266 3e-71
TC218822 183 2e-46
TC205752 178 8e-45
TC225718 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6... 172 2e-44
TC205753 172 3e-43
TC211182 similar to GB|AAO63406.1|28950965|BT005342 At1g03790 {A... 168 8e-42
TC224208 similar to GB|AAO63406.1|28950965|BT005342 At1g03790 {A... 99 8e-38
BG651885 similar to GP|8843784|dbj| zinc finger transcription fa... 152 5e-37
TC225716 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6... 130 2e-30
TC211980 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6... 124 2e-28
BM732973 similar to GP|14335106|gb| At2g41900/T6D20.20 {Arabidop... 122 4e-28
BG790659 113 3e-25
TC220991 weakly similar to GB|AAM16218.1|20334714|AY093957 At2g4... 100 2e-21
AW568218 96 5e-20
AW508155 96 5e-20
>TC224861 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription factor-like
protein (AT5g58620/mzn1_70), partial (42%)
Length = 2560
Score = 1023 bits (2646), Expect = 0.0
Identities = 535/694 (77%), Positives = 593/694 (85%), Gaps = 30/694 (4%)
Frame = +1
Query: 1 MCSGSKSKLSSSEQV-MES---QKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVN 56
MCS SKSKLSS V ME+ QK+N DGL+N S+LLELSAS+D+EA KREV+EKGLDVN
Sbjct: 106 MCSDSKSKLSSPTLVVMENSNIQKQNLDGLYN-SVLLELSASDDYEAFKREVEEKGLDVN 282
Query: 57 EAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLV--ETGRVDVNKACGSDKATAL 114
EAGFWYGRRIGSKKMGSE RTPLMIASLFGS +V+ Y++ + G VDVN+ CGSD+ATAL
Sbjct: 283 EAGFWYGRRIGSKKMGSETRTPLMIASLFGSAKVLNYILLQKGGGVDVNRVCGSDRATAL 462
Query: 115 HCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRG 174
HCAVAGGSE SLE+VK+LLDAG+DA+ LDA GNKP+NLIAPAF+S SKSRRKA+E+ LRG
Sbjct: 463 HCAVAGGSESSLEIVKLLLDAGADAECLDASGNKPVNLIAPAFDSLSKSRRKALEMFLRG 642
Query: 175 G-ESGQLIHQEMDLELISAPFSSKEGSD-KKEYPVDVSLPDINNGLYGSDEFRMYSFKVK 232
G E +L+ QEM+L++ S P KEGSD KKEYPVD+SLPDINNG+YG+DEFRMY+FKVK
Sbjct: 643 GGERDELMSQEMELQMFSVP-EKKEGSDNKKEYPVDISLPDINNGVYGTDEFRMYNFKVK 819
Query: 233 PCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFES 292
PCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKG CQKGDSCEYAHGVFES
Sbjct: 820 PCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGTCQKGDSCEYAHGVFES 999
Query: 293 WLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMS 352
WLHPAQYRTRLCKDETGC RKVCFFAH+PEELRPVYASTGSAMPSPKSYS++ +DM+ MS
Sbjct: 1000WLHPAQYRTRLCKDETGCARKVCFFAHKPEELRPVYASTGSAMPSPKSYSASGLDMTAMS 1179
Query: 353 PLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSA 412
PLALSS+SLPM TVSTPPMSPLA + SPKSG++WQNKI NLTPPSLQLPGSRLK ALSA
Sbjct: 1180PLALSSTSLPMPTVSTPPMSPLAAASSPKSGSMWQNKI--NLTPPSLQLPGSRLKAALSA 1353
Query: 413 RDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFR----NRIGDLTPTNLDDLL 468
RD ++EME+LGL SP R QQQQQQQQLIEEIARISSPSFR NRI DL PTNLDDLL
Sbjct: 1354RDLEMEMELLGLESPAR--QQQQQQQQLIEEIARISSPSFRSKEFNRIVDLNPTNLDDLL 1527
Query: 469 GSVDSNLLSQLHGLS-GPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLG 527
S D ++ SQLHGLS PSTPT Q M+QNMNHLR+SYPSNIPSSPVRKPS+ G
Sbjct: 1528ASADPSVFSQLHGLSVQPSTPTQSGLQ-----MRQNMNHLRASYPSNIPSSPVRKPSAFG 1692
Query: 528 FDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPS 587
FDSS+AVA AVMNSRSAAFAKRSQSFIDRGAA THHLG+SS SN SCRVSS SDW+SP+
Sbjct: 1693FDSSAAVATAVMNSRSAAFAKRSQSFIDRGAA-THHLGLSSASNSSCRVSSTLSDWSSPT 1869
Query: 588 GKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEH-AEPDVSWVHSLVKD----- 641
GKLDWGVNGD+L+KLRKS SFGFRN+G+ T SP+A E AEPDVSWVHSLVKD
Sbjct: 1870GKLDWGVNGDKLNKLRKSTSFGFRNSGV--TASPIAQPEFGAEPDVSWVHSLVKDVPSER 2043
Query: 642 -----------DLSKEMLPPWVEQLYIEQEQMVA 664
DLSKEMLPPW+EQLYIEQEQMVA
Sbjct: 2044SEIFGAEKQQYDLSKEMLPPWMEQLYIEQEQMVA 2145
>TC224880 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription factor-like
protein (AT5g58620/mzn1_70), partial (21%)
Length = 1723
Score = 598 bits (1542), Expect = e-171
Identities = 321/426 (75%), Positives = 345/426 (80%), Gaps = 24/426 (5%)
Frame = +3
Query: 252 ENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCG 311
ENARRRDPRKYPYSCVPCPEFRKG CQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGC
Sbjct: 9 ENARRRDPRKYPYSCVPCPEFRKGTCQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCA 188
Query: 312 RKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPM 371
RKVCFFAH+PEELRPVYASTGSAMPSPKSYS++ +DM+ MSPLALSS+SLPM TVSTPPM
Sbjct: 189 RKVCFFAHKPEELRPVYASTGSAMPSPKSYSASGLDMTAMSPLALSSTSLPMPTVSTPPM 368
Query: 372 SPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTR-Q 430
SPL S SPKSG+LWQNKI NLTPPSLQLPGSRLK ALSARD ++EME+LGL SP R
Sbjct: 369 SPLTAS-SPKSGSLWQNKI--NLTPPSLQLPGSRLKAALSARDLEMEMELLGLESPARHH 539
Query: 431 QQQQQQQQQLIEEIARISSPSFR----NRIGDLTPTNLDD-LLGSVDSNLLSQLHGLSGP 485
QQQQQQQLIEEIARISSPSFR NRIGDL PTNLDD LL S D ++LSQL
Sbjct: 540 HHQQQQQQQLIEEIARISSPSFRSKEFNRIGDLNPTNLDDLLLASADPSVLSQL------ 701
Query: 486 STPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAA 545
QS QM Q NHLR+SYPSN+PSSPVRKPSS GFDSS+AVA A+MNSRSAA
Sbjct: 702 --------QSGLQMRQSMNNHLRASYPSNVPSSPVRKPSSFGFDSSAAVATAMMNSRSAA 857
Query: 546 FAKRSQSFIDRGAAGTHHL--GMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLR 603
FAKRSQSFIDRGAA THH GMSSPSNPSCRVSS S W+SP+GKLDWGVNGDEL+KLR
Sbjct: 858 FAKRSQSFIDRGAAATHHHLGGMSSPSNPSCRVSSTLSGWSSPTGKLDWGVNGDELNKLR 1037
Query: 604 KSASFGFRNNGM--APTGSPMAHSEH-AEPDVSWVHSLVK------------DDLSKEM- 647
KSASFGFRN+G+ + + SP+A E E DVSWVHSLVK D LSKEM
Sbjct: 1038KSASFGFRNSGVTASSSSSPIAQPEFGTEQDVSWVHSLVKDVPSERSEKQQYDHLSKEML 1217
Query: 648 LPPWVE 653
L PWVE
Sbjct: 1218LSPWVE 1235
Score = 37.7 bits (86), Expect = 0.017
Identities = 19/22 (86%), Positives = 20/22 (90%), Gaps = 1/22 (4%)
Frame = +2
Query: 644 SKEML-PPWVEQLYIEQEQMVA 664
SKEML PWVEQLYIEQ+QMVA
Sbjct: 1256 SKEMLLSPWVEQLYIEQKQMVA 1321
>TC206602 similar to UP|Q9LUZ4 (Q9LUZ4) Zinc finger transcription factor-like
protein (AT5g58620/mzn1_70), partial (31%)
Length = 1855
Score = 510 bits (1313), Expect = e-145
Identities = 268/508 (52%), Positives = 347/508 (67%), Gaps = 46/508 (9%)
Frame = +1
Query: 203 KEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKY 262
K+YP+D+SLPDI NG+YG+DEFRMY+FKVKPCSRAYSHDWTECPFVHPGENARRRDPRKY
Sbjct: 19 KDYPIDLSLPDIKNGIYGTDEFRMYTFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKY 198
Query: 263 PYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPE 322
YSCVPCPEFRKG+C KGD+CEYAHG+FE WLHPAQYRTRLCKDE GC R+VCFFAH+ E
Sbjct: 199 HYSCVPCPEFRKGSCSKGDACEYAHGIFECWLHPAQYRTRLCKDEGGCTRRVCFFAHKLE 378
Query: 323 ELRPVYASTGSAMPSPKSY--SSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSP 380
ELRP+YASTGSA+PSP+SY S++A++M ++P+AL S S+ M STPP++P +G+ SP
Sbjct: 379 ELRPLYASTGSAIPSPRSYSASASALEMGSVNPIALGSPSVLMPPTSTPPLTP-SGASSP 555
Query: 381 KSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQL 440
+G++W N++ P+LQLP SRLKTA + RD DL+ME+LGL + ++QQ +
Sbjct: 556 IAGSMWSQS---NVSVPTLQLPKSRLKTASTVRDTDLDMELLGL------ETHWRRQQLM 708
Query: 441 IEEIARISSPSFR--------------------NRIGDLTPTNLDDLLGSVDSNLLSQLH 480
++EI+ +SSP+++ NR+ + P NL+D+ GS+D ++LS+ H
Sbjct: 709 MDEISALSSPNWKNSMPNSPSFRVPLNDHTGELNRLSGVKPANLEDMFGSLDPSILSKYH 888
Query: 481 GLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMN 540
G+S LQS + Q M+QN+N Y S++ + V S D S A+ +N
Sbjct: 889 GISLDVAGPQLQSPTGIQ-MRQNVNQQLGGYSSSLSTLNVIGSRSFRLDQSGEAASVALN 1065
Query: 541 SRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELS 600
R AAFAKRSQSFI+RG HH + SP S FS+W SP GKLDW VNG+EL+
Sbjct: 1066PRVAAFAKRSQSFIERGVV-NHHSELPSPK------PSTFSNWGSPVGKLDWAVNGEELN 1224
Query: 601 KLRKSASFGFRNNG--MAPTGSPMAHSEHAEPDVSWVHSLVKD--------------DLS 644
KLRKSASFGFR + + T + M+ + EPDVSWV+SLVKD D
Sbjct: 1225KLRKSASFGFRGSDTPLTKTSTKMSANVDDEPDVSWVNSLVKDAPPESGESGEYSVEDQR 1404
Query: 645 K--------EMLPPWVEQLYIEQEQMVA 664
K + +P W+EQLY++QEQMVA
Sbjct: 1405KLLQCHNGTDAIPAWLEQLYLDQEQMVA 1488
>TC225715 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (42%)
Length = 2142
Score = 357 bits (915), Expect = 1e-98
Identities = 209/459 (45%), Positives = 277/459 (59%), Gaps = 17/459 (3%)
Frame = +3
Query: 200 SDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDP 259
S+KKEYP+D SLPDI N +Y +DEFRM+SFKV+PCSRAYSHDWTECPFVHPGENARRRDP
Sbjct: 9 SEKKEYPIDPSLPDIKNSIYATDEFRMFSFKVRPCSRAYSHDWTECPFVHPGENARRRDP 188
Query: 260 RKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAH 319
RK+ YSCVPCP+FRKGAC++GD CEYAHGVFE WLHPAQYRTRLCKD T C R+VCFFAH
Sbjct: 189 RKFHYSCVPCPDFRKGACRRGDMCEYAHGVFECWLHPAQYRTRLCKDGTSCNRRVCFFAH 368
Query: 320 RPEELRPVYASTGSAMPSPKSYSS--NAVDMSV---MSPLALSSSSLPMSTVSTPPMSPL 374
EELRP+Y STGSA PSP+S +S N +DM+ + P + SS S + PMSP
Sbjct: 369 TAEELRPLYVSTGSAAPSPRSSASGPNVMDMAAAMSLFPGSPSSGSSMSPSHFGQPMSPS 548
Query: 375 AGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQ 434
A + S N L+L P L SRL+++LSARD E + +QQ
Sbjct: 549 ANGMPLSSAWAQPNVPALHL--PGSNLQSSRLRSSLSARDIPPEDLNMMSDLDGQQQHHL 722
Query: 435 QQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPS--TPTHLQ 492
I+ SS S R LTP+NL++L S + +L + + S +PTH
Sbjct: 723 NDLSCYIQPRPGASSVSRSGRSKTLTPSNLEELF-SAEISLSPRYSDPAAGSVFSPTHKS 899
Query: 493 S-QSRHQMMQQNMNHLRSSY--PSNIPSSPVRKPSSLGFDSS------SAVAAAVMNSRS 543
+ ++ Q +Q ++ + ++ P N+ + +S G S S + M++R
Sbjct: 900 AVLNQFQQLQSMLSPINTNLLSPKNVEHPLFQ--ASFGVSPSGRMSPRSVEPISPMSARL 1073
Query: 544 AAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSK-L 602
+AFA+R + + + LG +SP++ ++ +S W SP GK DW VNGD L + +
Sbjct: 1074SAFAQREKQQQQLRSVSSRDLGANSPASLVGSPANPWSKWGSPIGKADWSVNGDSLGRQM 1253
Query: 603 RKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD 641
R+S+SF +NNG EPD+SWV SLVK+
Sbjct: 1254RRSSSFERKNNG-------------EEPDLSWVQSLVKE 1331
>TC231961 homologue to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (25%)
Length = 893
Score = 322 bits (826), Expect = 3e-88
Identities = 161/275 (58%), Positives = 199/275 (71%), Gaps = 9/275 (3%)
Frame = +2
Query: 196 SKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENAR 255
+ E S+KKEYPVD+SLPDI N +Y SDEFRMYSFKV+PCSRAYSHDWTECPFVHPGENAR
Sbjct: 104 TNEVSEKKEYPVDLSLPDIKNSIYSSDEFRMYSFKVRPCSRAYSHDWTECPFVHPGENAR 283
Query: 256 RRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVC 315
RRDPRK+ YSCVPCPEFRKGAC++GD CEYAHGVFE WLHPAQYRTRLCKD T C R+VC
Sbjct: 284 RRDPRKFHYSCVPCPEFRKGACRRGDMCEYAHGVFECWLHPAQYRTRLCKDGTNCARRVC 463
Query: 316 FFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLA 375
FFAH EELRP+Y STGSA+PSP+S +S+A+D ++ ++ SS S+ + TPPMSP +
Sbjct: 464 FFAHTNEELRPLYVSTGSAVPSPRSSASSAMDF--VAAISPSSMSVMSPSPFTPPMSPSS 637
Query: 376 GSLSPKSGNLWQNKINLNLTPPSLQLPG-----SRLKTALSARDFDLEMEMLGLGSPTRQ 430
S++ N+ P+L LPG SRL+++L+ARDF ++ L L
Sbjct: 638 ASIAWPQPNI-----------PALHLPGSNFHSSRLRSSLNARDFSVDDFDLLLPDYDHH 784
Query: 431 QQQQQQQQQLIEEIARISSPSFR----NRIGDLTP 461
QQQQQQQ + E++ +S + NR G + P
Sbjct: 785 HHQQQQQQQFLNELSCLSPHAMNCNTMNRSGRMKP 889
>BQ295977 similar to GP|8843784|dbj zinc finger transcription factor-like
protein {Arabidopsis thaliana}, partial (22%)
Length = 422
Score = 266 bits (679), Expect = 3e-71
Identities = 115/137 (83%), Positives = 128/137 (92%)
Frame = +3
Query: 206 PVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYS 265
PVD+SLPDI NG+Y SDEFRMY+FKV+PCSRAYSHDWTECPFVHPGENARRRDPR+Y YS
Sbjct: 3 PVDLSLPDIKNGIYSSDEFRMYTFKVRPCSRAYSHDWTECPFVHPGENARRRDPRRYQYS 182
Query: 266 CVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELR 325
CVPCPEFRKG+C KGD+C+YAHG+FE WLHPAQY+TRLCK ETGC R+VCFFAH E+LR
Sbjct: 183 CVPCPEFRKGSCSKGDACDYAHGIFECWLHPAQYKTRLCK-ETGCTRRVCFFAHNVEDLR 359
Query: 326 PVYASTGSAMPSPKSYS 342
PVYASTGSAMPSP+SYS
Sbjct: 360 PVYASTGSAMPSPRSYS 410
>TC218822
Length = 554
Score = 183 bits (464), Expect = 2e-46
Identities = 83/138 (60%), Positives = 97/138 (70%)
Frame = +3
Query: 188 ELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPF 247
E A S+ S + E PVD Y D FRM+ FKV+ C+R SHDWT+CP+
Sbjct: 168 ESTDAESDSEVPSREPEVPVDA---------YSCDHFRMFEFKVRRCARCRSHDWTDCPY 320
Query: 248 VHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDE 307
HPGE ARRRDPRKY YS CP+FRKG+C+KGD+CEYAHGVFE WLHPA+YRT+ CKD
Sbjct: 321 AHPGEKARRRDPRKYHYSGTACPDFRKGSCKKGDACEYAHGVFECWLHPARYRTQPCKDG 500
Query: 308 TGCGRKVCFFAHRPEELR 325
T C R VCFFAH PE+LR
Sbjct: 501 TSCRRCVCFFAHTPEQLR 554
>TC205752
Length = 1020
Score = 178 bits (451), Expect = 8e-45
Identities = 77/129 (59%), Positives = 98/129 (75%), Gaps = 1/129 (0%)
Frame = +1
Query: 218 LYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGAC 277
L+ SD FRM+ FKV+ C R SHDWTECP+ HP E ARRRDPRKY YS CP++RKG C
Sbjct: 91 LFSSDHFRMFQFKVRNCPRGRSHDWTECPYAHPAEKARRRDPRKYHYSGTSCPDYRKGNC 270
Query: 278 QKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVY-ASTGSAMP 336
++GD+C++AHGVFE WLHP++YRT+LCKD T C R+VCFFAH ++LR V +S+ S +
Sbjct: 271 KRGDTCQFAHGVFECWLHPSRYRTQLCKDGTNCRRRVCFFAHTSDQLRVVTDSSSSSFVS 450
Query: 337 SPKSYSSNA 345
SP S N+
Sbjct: 451 SPTSVLINS 477
>TC225718 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (38%)
Length = 1643
Score = 172 bits (435), Expect(2) = 2e-44
Identities = 128/344 (37%), Positives = 184/344 (53%), Gaps = 19/344 (5%)
Frame = +2
Query: 290 FESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSS--NAVD 347
FE WLHPAQYRTRLCKD T C R+VCFFAH EELRP+Y STGSA+PSP+S +S N +D
Sbjct: 2 FECWLHPAQYRTRLCKDGTSCNRRVCFFAHTAEELRPLYVSTGSAVPSPRSSASAPNVMD 181
Query: 348 M-SVMSPLALSSSSLPMSTVS--TPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGS 404
M + MS L S SS+ + S PMSP A +S S N L+L P L S
Sbjct: 182 MAAAMSLLPGSPSSVSSMSPSHFGQPMSPSANGMSLSSAWAQPNVSALHL--PGSNLQSS 355
Query: 405 RLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNL 464
RL+++LSARD + + +QQ ++ S S R LTP+NL
Sbjct: 356 RLRSSLSARDMPPDDLNMMSDLDGQQQHPLNDLSCYLQPRPGAGSVSRSGRSKILTPSNL 535
Query: 465 DDLLGSVDSNLLSQLHGLSGP-STPTHLQS-QSRHQMMQQNMNHLRSSY--PSNIPSSPV 520
+DL + S+ +G +PTH + ++ Q +Q ++ + ++ P N+ +
Sbjct: 536 EDLFSAEISSSPRYSDPAAGSVFSPTHKSAVLNQFQQLQSMLSPINTNLLSPKNVEHPLL 715
Query: 521 RKPSSLGFDSS------SAVAAAVMNSRSAAFA---KRSQSFIDRGAAGTHHLGMSSPSN 571
+ +S G S S + M+SR +AFA K+ Q + + LG +SP++
Sbjct: 716 Q--ASFGVSPSGRMSPRSVEPISPMSSRISAFAQREKQQQQQQQLRSLSSRDLGANSPAS 889
Query: 572 PSCRVSSGFSDWNSPSGKLDWGVNGDELSK-LRKSASFGFRNNG 614
++ +S W SP+GK DW VNGD L + +R+S+SF +NNG
Sbjct: 890 LVGSPANPWSKWGSPNGKADWSVNGDTLGRQMRRSSSFELKNNG 1021
Score = 26.6 bits (57), Expect(2) = 2e-44
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = +3
Query: 629 EPDVSWVHSLVKD 641
EPD+SWV SLVK+
Sbjct: 1026 EPDLSWVQSLVKE 1064
>TC205753
Length = 457
Score = 172 bits (437), Expect = 3e-43
Identities = 71/117 (60%), Positives = 90/117 (76%)
Frame = +3
Query: 218 LYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGAC 277
L+ SD FRM+ FKV+ C R SHDWTECP+ HP E ARRRDPRKY YS CP+++KG C
Sbjct: 105 LFSSDHFRMFQFKVRICPRGRSHDWTECPYAHPAEKARRRDPRKYHYSGTACPDYQKGNC 284
Query: 278 QKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSA 334
++GD+C+++HGVFE WLHP++YRT LCKD T C R+VCFFAH E+LR V + S+
Sbjct: 285 KRGDTCQFSHGVFECWLHPSRYRTHLCKDGTTCRRRVCFFAHTTEQLRLVVTDSSSS 455
>TC211182 similar to GB|AAO63406.1|28950965|BT005342 At1g03790 {Arabidopsis
thaliana;} , partial (27%)
Length = 700
Score = 168 bits (425), Expect = 8e-42
Identities = 71/111 (63%), Positives = 87/111 (77%), Gaps = 1/111 (0%)
Frame = +2
Query: 208 DVSLPDINNG-LYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSC 266
D S D ++G Y SD+FRM+ FKV+ CSR+ SHDWT+CPFVHPGE ARRRDPR++ YS
Sbjct: 236 DSSTDDDSDGDPYASDQFRMFEFKVRRCSRSRSHDWTDCPFVHPGEKARRRDPRRFHYSA 415
Query: 267 VPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFF 317
CPEFR+G C +GD+CE++HGVFE WLHP++YRT CKD C RKVCFF
Sbjct: 416 TVCPEFRRGQCDRGDACEFSHGVFECWLHPSRYRTEACKDGKNCKRKVCFF 568
>TC224208 similar to GB|AAO63406.1|28950965|BT005342 At1g03790 {Arabidopsis
thaliana;} , partial (28%)
Length = 884
Score = 99.4 bits (246), Expect(2) = 8e-38
Identities = 39/59 (66%), Positives = 47/59 (79%)
Frame = +2
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGAC 277
Y SD FRM+ FKV+ C+R+ SHDWT+CPF HPGE ARRRDPR+Y YS CPE+R+G C
Sbjct: 281 YSSDHFRMFEFKVRQCTRSRSHDWTDCPFAHPGEKARRRDPRRYHYSGTVCPEYRRGGC 457
Score = 77.0 bits (188), Expect(2) = 8e-38
Identities = 35/61 (57%), Positives = 40/61 (65%)
Frame = +3
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
+ D+CEYAHGVFE WLHP +YRT CKD C RKVCFFAH P + V TG +P
Sbjct: 462 RDDACEYAHGVFECWLHPFRYRTEACKDGRNCKRKVCFFAHTPXQ---VENFTGXILPFX 632
Query: 339 K 339
K
Sbjct: 633 K 635
>BG651885 similar to GP|8843784|dbj| zinc finger transcription factor-like
protein {Arabidopsis thaliana}, partial (3%)
Length = 389
Score = 152 bits (384), Expect = 5e-37
Identities = 93/169 (55%), Positives = 100/169 (59%), Gaps = 17/169 (10%)
Frame = +2
Query: 501 QQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAG 560
+QNMNHLR+SYPSNIPSSPVRKPS+ GFDSS+A A A MNSRSAAFAKRSQSFIDRGAA
Sbjct: 8 RQNMNHLRASYPSNIPSSPVRKPSAFGFDSSAAAATAGMNSRSAAFAKRSQSFIDRGAA- 184
Query: 561 THHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGS 620
THHLG+SS SN SCRV T S
Sbjct: 185 THHLGLSSASNSSCRV-----------------------------------------TAS 241
Query: 621 PMAHSEH-AEPDVSWVHSLVKD----------------DLSKEMLPPWV 652
P+A E AEPDVSWVHSLVKD DLSKEMLPPW+
Sbjct: 242 PIAQPEFGAEPDVSWVHSLVKDVPSERSEIFGAEKQQYDLSKEMLPPWM 388
>TC225716 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (18%)
Length = 1644
Score = 130 bits (326), Expect = 2e-30
Identities = 113/352 (32%), Positives = 173/352 (49%), Gaps = 30/352 (8%)
Frame = +1
Query: 324 LRPVYASTGSAMPSPKSYSS--NAVDMSVMSPLALSSSSLPMSTVSTP---PMSPLAGSL 378
LRP+Y STGSA+PSP+S +S N +DM+ L S S S +P PMSP +
Sbjct: 7 LRPLYVSTGSAVPSPRSSASTPNVMDMAAAMSLFPGSPSSISSMSPSPFVQPMSPSTSGI 186
Query: 379 SPKSGNLWQNKINLNLTPPSLQLPGS-----RLKTALSARDFDLE-MEMLGLGSPTRQQQ 432
S S N W P+L LPGS RL+++LSARD E ++L Q
Sbjct: 187 S-HSSNAWPQP-----NVPALHLPGSNIQTSRLRSSLSARDMPPEDFDVL---------Q 321
Query: 433 QQQQQQQLIEEIARISSP-------SFRNRIGDLTPTNLDDLLGS-VDSNLLSQLHGLSG 484
QQ L+ ++ S P S R LTP+NLD+L + + S+ ++
Sbjct: 322 DFDGQQHLLSDLGCFSQPRPGAISVSRSGRSKTLTPSNLDELFSAEISSSPRYSDPAVAS 501
Query: 485 PSTPTHLQS-QSRHQMMQQNMNHLRSSY--PSNIP------SSPVRKPSSLGFDSSSAVA 535
+P H + ++ Q +Q +++ + +S P N+ S V P + S ++
Sbjct: 502 VFSPRHKSTIMNQFQQLQSSLSPINTSVSSPRNVEHPLLQASFGVSSPGRMSPRSMEPIS 681
Query: 536 AAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSP--SGKLDWG 593
M+SR +AFA+R + + + LG + P++ + +S+W SP +GK+DW
Sbjct: 682 P--MSSRLSAFAQREKQHQQLRSLSSRDLGANVPASMVGSPVNSWSNWGSPHGNGKVDWS 855
Query: 594 VNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDLSK 645
VNG+EL +L++S+SF NNG EPD+SWV SLVK+ S+
Sbjct: 856 VNGNELGRLQRSSSFELGNNG-------------EEPDLSWVQSLVKESPSE 972
>TC211980 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (21%)
Length = 761
Score = 124 bits (310), Expect = 2e-28
Identities = 76/207 (36%), Positives = 122/207 (58%), Gaps = 27/207 (13%)
Frame = +3
Query: 27 HNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFG 86
++ S LLEL+A+ND KR ++ ++E G WY R SKKM +E+RTPLM+A+ +G
Sbjct: 108 YSLSSLLELAANNDVSGFKRLIECDPSSIDEVGLWYIRHKESKKMVNEQRTPLMVAATYG 287
Query: 87 STRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACG 146
S V+K ++ DVN +CG DK+TALHCA +GGSE +++ VK+LL+AG+D +S+D
Sbjct: 288 SIDVMKLILSLSEADVNLSCGLDKSTALHCAASGGSENAVDAVKLLLEAGADVNSVDVNA 467
Query: 147 NKPMNLIAPAFNSSSKSRRK-AMELLLRGGESGQLIH------------------QEMDL 187
++P ++I F + + +K ++E LL+ + L+ E+++
Sbjct: 468 HRPGDVI--VFPTKLEHVKKTSLEELLQKTDDWSLLRVITTTTSCNACSPPLSTSPEIEI 641
Query: 188 EL-ISAPFS-------SKEGSDKKEYP 206
E+ I +P S S E ++KKEYP
Sbjct: 642 EIEIESPCSARDSKMKSDEVTEKKEYP 722
>BM732973 similar to GP|14335106|gb| At2g41900/T6D20.20 {Arabidopsis
thaliana}, partial (12%)
Length = 421
Score = 122 bits (307), Expect = 4e-28
Identities = 75/168 (44%), Positives = 96/168 (56%), Gaps = 5/168 (2%)
Frame = +2
Query: 286 AHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNA 345
AHGVFE WLHPAQYRTRLCKD T C R+VCFFAH EELRP+Y STGSA+PSP+S
Sbjct: 2 AHGVFECWLHPAQYRTRLCKDGTNCARRVCFFAHTNEELRPLYVSTGSAVPSPRS----- 166
Query: 346 VDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPG-- 403
+ S M V+ MSP + S++ N+ P+L LPG
Sbjct: 167 -----------GAPSSAMDFVTAMTMSPSSPSIAWSQPNI-----------PALHLPGSN 280
Query: 404 ---SRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARIS 448
SRL+++L+AR D+ M+ L P Q QQQQQ + E++ +S
Sbjct: 281 FHSSRLRSSLNAR--DISMDDFDLLLP--DYDQHQQQQQFLNELSCLS 412
>BG790659
Length = 358
Score = 113 bits (282), Expect = 3e-25
Identities = 55/63 (87%), Positives = 58/63 (91%)
Frame = +1
Query: 24 DGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIAS 83
DGL+N S+LLELSA +DFEA KREVDEKGLDVNEAG WYGRRIGSKKMGSE RTPLMIAS
Sbjct: 169 DGLYNNSVLLELSAXDDFEAFKREVDEKGLDVNEAGLWYGRRIGSKKMGSETRTPLMIAS 348
Query: 84 LFG 86
LFG
Sbjct: 349 LFG 357
>TC220991 weakly similar to GB|AAM16218.1|20334714|AY093957
At2g41900/T6D20.20 {Arabidopsis thaliana;} , partial
(14%)
Length = 681
Score = 100 bits (249), Expect = 2e-21
Identities = 51/115 (44%), Positives = 77/115 (66%)
Frame = +1
Query: 8 KLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIG 67
K+++++ M+S N + +FS LLEL+++ND E K +++ +NE G WYGR+ G
Sbjct: 337 KVTANDIEMKSLTVNTED--SFSSLLELASNNDIEGFKVLLEKDSSTINEVGLWYGRQNG 510
Query: 68 SKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGS 122
SK+ E RTPLM+A+ +GS V+K ++ DVN ACG++K TALHCA +GGS
Sbjct: 511 SKQFVLEHRTPLMVAATYGSIDVMKMVLLCPEADVNFACGANKTTALHCAASGGS 675
>AW568218
Length = 427
Score = 95.9 bits (237), Expect = 5e-20
Identities = 40/64 (62%), Positives = 49/64 (76%)
Frame = +3
Query: 210 SLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPC 269
S D + Y SD+FRM+ FKV+ C+R+ SHDWT+CPFVHPGE ARRRDPR++ YS C
Sbjct: 234 SSDDDSGDPYASDQFRMFEFKVRRCTRSRSHDWTDCPFVHPGEKARRRDPRRFHYSATVC 413
Query: 270 PEFR 273
PEFR
Sbjct: 414 PEFR 425
>AW508155
Length = 220
Score = 95.9 bits (237), Expect = 5e-20
Identities = 39/69 (56%), Positives = 51/69 (73%)
Frame = +2
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
Y SD+FRM+ FKV+ +R+ SHDWT+ PFVH GENARRRDPR+ Y CPEF + C
Sbjct: 14 YASDQFRMFEFKVRRLTRSRSHDWTD*PFVHTGENARRRDPRRIQYYATGCPEFLRSQCD 193
Query: 279 KGDSCEYAH 287
+G++CE +H
Sbjct: 194 RGNACEISH 220
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,568,342
Number of Sequences: 63676
Number of extensions: 483917
Number of successful extensions: 3634
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 2941
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3294
length of query: 664
length of database: 12,639,632
effective HSP length: 103
effective length of query: 561
effective length of database: 6,081,004
effective search space: 3411443244
effective search space used: 3411443244
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0204.2


