

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
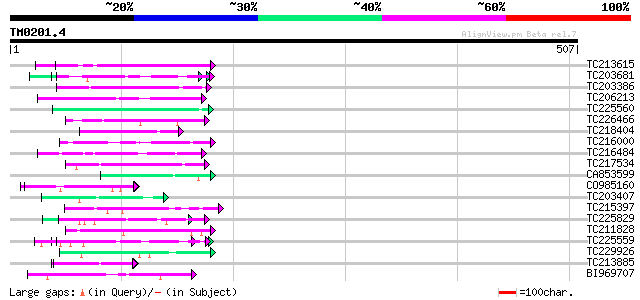
Query= TM0201.4
(507 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase ... 54 2e-07
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 51 1e-06
TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 48 9e-06
TC206213 similar to UP|PMXA_MOUSE (Q62066) Paired mesoderm homeo... 48 1e-05
TC225560 similar to UP|Q9FUL8 (Q9FUL8) Arabinogalactan protein, ... 47 2e-05
TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {... 47 2e-05
TC218404 UP|Q9MAX7 (Q9MAX7) Epsilon2-COP (Fragment), partial (49%) 47 2e-05
TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light rec... 47 3e-05
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 46 3e-05
TC217534 similar to GB|AAB62826.1|2252827|IG005I10 coded for by ... 46 4e-05
CA853599 weakly similar to PIR|F86379|F86 protein F21J9.28 [impo... 46 4e-05
CO985160 46 4e-05
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 46 4e-05
TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_4... 45 6e-05
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 45 6e-05
TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-pho... 45 6e-05
TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 45 8e-05
TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, part... 45 1e-04
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 44 1e-04
BI969707 44 2e-04
>TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase
(CBL-interacting protein kinase 4), partial (45%)
Length = 674
Score = 53.9 bits (128), Expect = 2e-07
Identities = 37/143 (25%), Positives = 66/143 (45%)
Frame = +3
Query: 42 AGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAA 101
A T+ P + +T ++P S +++T P A ASS S+ A AA + S + +
Sbjct: 258 ASTTTPTSSKSTRSSPPRPR----STSSSTSPVA--ASSSPSSPAAAASRSPSPAATSRS 419
Query: 102 SASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCP 161
S+ ++A+ A +S T + TP + R SPPS + + ++ S P + P
Sbjct: 420 SSPLSASATAMASPTETSSHRTSSSTPPATSRSPTSASPPSRNTSTTVSSTPPAARRPSP 599
Query: 162 GAATTSEAAQIEQAPAPEVGSSS 184
+++ +A P P +SS
Sbjct: 600 RQRSSAASATTAXKPTPGPAASS 668
Score = 42.4 bits (98), Expect = 5e-04
Identities = 41/161 (25%), Positives = 67/161 (41%)
Frame = +3
Query: 24 AESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPAS 83
A ST P + T++S S + AA+ + A ++ + + +SSP S
Sbjct: 255 AASTTTPTSSKSTRSSPPRPRSTSSSTSPVAASSSPSSPAAAASRSPSPAATSRSSSPLS 434
Query: 84 AGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPST 143
A ATA AS + ++ +SS P + + T SPP ++ + ST
Sbjct: 435 ASATA-----------MASPTETSSHRTSSSTPPATSR---SPTSASPPSRNTSTTVSST 572
Query: 144 HDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 184
A PSP ++S +ATT+ + P P SS+
Sbjct: 573 PPAARRPSP---RQRSSAASATTA----XKPTPGPAASSST 674
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 51.2 bits (121), Expect = 1e-06
Identities = 44/141 (31%), Positives = 61/141 (43%)
Frame = +3
Query: 43 GTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAAS 102
G +P A PTT S AA T P A PA++P+ + A VA+ +S A+S
Sbjct: 69 GGQSPAASPTT-----------SPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASS 215
Query: 103 ASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPG 162
NA A + +P T SPP + A P+P +T A + P+ + P
Sbjct: 216 P--NAATATPPASSP---------TVASPPSKAAAPAPVATPPAATPPA-------ATPP 341
Query: 163 AATTSEAAQIEQAPAPEVGSS 183
AAT + PAP SS
Sbjct: 342 AATPPAVTPVSSPPAPVPVSS 404
Score = 44.7 bits (104), Expect = 1e-04
Identities = 46/163 (28%), Positives = 61/163 (37%)
Frame = +3
Query: 18 GGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVP 77
GG A T +P T +S A + P A+P + SS AAT P
Sbjct: 69 GGQSPAASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAAT--ATPP 242
Query: 78 ASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAP 137
ASSP A + AA + + A+ AT AA+ PP
Sbjct: 243 ASSPTVASPPSKAAAPAPVATPPAATPPAATPPAAT-----------------PPAVTPV 371
Query: 138 PSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEV 180
SPP+ S P+P S P A T+ A + AP+ EV
Sbjct: 372 SSPPAPVPVSSPPAP--VPVSSPPALAPTTPAPVV--APSAEV 488
Score = 43.9 bits (102), Expect = 2e-04
Identities = 39/151 (25%), Positives = 61/151 (39%), Gaps = 15/151 (9%)
Frame = +3
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVA-------------AATEPTAVPASSPASA 84
+S +A T+ P A T A+P K A + VA AAT P P SSP +
Sbjct: 210 SSPNAATATPPASSPTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAP 389
Query: 85 GATAAVAAESSIGATAASA-SVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPST 143
++ A + + A A + A A S++ P + + +T KS PSP
Sbjct: 390 VPVSSPPAPVPVSSPPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKSKKHTAPAPSPSLL 569
Query: 144 -HDAGSMPSPPHQGEKSCPGAATTSEAAQIE 173
A + +P + PG A + + + E
Sbjct: 570 GPPAPPVGAPGSSQDSMSPGPAVSEDESGAE 662
>TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 649
Score = 48.1 bits (113), Expect = 9e-06
Identities = 39/138 (28%), Positives = 56/138 (40%)
Frame = +1
Query: 43 GTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAAS 102
G +P A PTT++A ++ + P A P SSP ++ AA A T AS
Sbjct: 64 GGQSPAAAPTTSSASPATAPSKPKPKSPA-PVASPTSSPPASSPNAATATPPVSSPTVAS 240
Query: 103 ASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPG 162
A A ++ P+ +PP SPP+ S P+P S P
Sbjct: 241 PPSKAAAPAPATTPPVATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAP--VPVSSPPA 414
Query: 163 AATTSEAAQIEQAPAPEV 180
A T+ A + AP+ EV
Sbjct: 415 LAPTTPAPVV--APSAEV 462
Score = 40.8 bits (94), Expect = 0.001
Identities = 37/151 (24%), Positives = 59/151 (38%), Gaps = 15/151 (9%)
Frame = +1
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVA-------------AATEPTAVPASSPASA 84
+S +A T+ P T A+P K A + AAT P P SSP +
Sbjct: 184 SSPNAATATPPVSSPTVASPPSKAAAPAPATTPPVATPPAATPPAATPPAVTPVSSPPAP 363
Query: 85 GATAAVAAESSIGATAASA-SVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPST 143
++ A + + A A + A A S++ P + + +T KS PSP
Sbjct: 364 VPVSSPPAPVPVSSPPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKSKKHTAPAPSPALL 543
Query: 144 -HDAGSMPSPPHQGEKSCPGAATTSEAAQIE 173
A + +P + S PG A + + + E
Sbjct: 544 GPPAPPVGAPGSSQDASSPGPAVSEDESGAE 636
Score = 39.3 bits (90), Expect = 0.004
Identities = 34/120 (28%), Positives = 49/120 (40%)
Frame = +1
Query: 64 ESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEK 123
+S AA T +A PA++P+ + S + AS S NA A +P
Sbjct: 70 QSPAAAPTTSSASPATAPSKPKPKSPAPVASPTSSPPAS-SPNAATATPPVSSP------ 228
Query: 124 ENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 183
T SPP + A P+P +T + P+ + P AAT + PAP SS
Sbjct: 229 ---TVASPPSKAAAPAPATTPPVATPPA-------ATPPAATPPAVTPVSSPPAPVPVSS 378
>TC206213 similar to UP|PMXA_MOUSE (Q62066) Paired mesoderm homeobox protein
2A (Paired-like homeobox 2A) (PHOX2A homeodomain
protein) (Aristaless homeobox protein homolog), partial
(8%)
Length = 930
Score = 47.8 bits (112), Expect = 1e-05
Identities = 43/151 (28%), Positives = 62/151 (40%)
Frame = +3
Query: 26 STKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAG 85
S P RR S G S + + + SS A + P+A A SPAS
Sbjct: 195 SAAPPWCRRSPPRSPPCGPSCGASTTRRPTSTSSRAATSSSATAMSGPSARSA*SPASLP 374
Query: 86 ATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHD 145
+A A+ SS T+A++S +A AA ++ T SP APP PS+
Sbjct: 375 PSAPSASTSS--TTSATSSASAWSAATTASP---------TTAPSPSSTPAPPPAPSSSS 521
Query: 146 AGSMPSPPHQGEKSCPGAATTSEAAQIEQAP 176
S S P ++ ++T S AA +P
Sbjct: 522 PTSSTSLPATPPRTLASSSTPSSAATSNPSP 614
Score = 32.7 bits (73), Expect = 0.39
Identities = 35/142 (24%), Positives = 52/142 (35%), Gaps = 2/142 (1%)
Frame = +3
Query: 45 SNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAS 104
+ P + P T+AAP + P + P A T + SS AT++SA+
Sbjct: 168 TTPMSCPRTSAAPPW--------CRRSPPRSPPCGPSCGASTTRRPTSTSSRAATSSSAT 323
Query: 105 VNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPS--THDAGSMPSPPHQGEKSCPG 162
+ +A S+ +P PPS PS T S S +
Sbjct: 324 AMSGPSARSA*SPAS----------------LPPSAPSASTSSTTSATSSASAWSAATTA 455
Query: 163 AATTSEAAQIEQAPAPEVGSSS 184
+ TT+ + AP P SSS
Sbjct: 456 SPTTAPSPSSTPAPPPAPSSSS 521
>TC225560 similar to UP|Q9FUL8 (Q9FUL8) Arabinogalactan protein, partial
(48%)
Length = 1030
Score = 47.0 bits (110), Expect = 2e-05
Identities = 41/145 (28%), Positives = 58/145 (39%), Gaps = 1/145 (0%)
Frame = +2
Query: 39 SSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGA 98
S A SN A P A + + S A+ ++ PASSP A AT A + +S
Sbjct: 137 SPAAAPSNTPATPAAATPAQAPSTPNSPAPVASPKSSPPASSPKPATATPASSPAASPTT 316
Query: 99 T-AASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGE 157
T AA A AA+ P+ P S P P + P+T A + H+ +
Sbjct: 317 TPAAPAPATKPPAASPPPAPVPVSSPPAPVPVSSPPAPVPVAAPTTPVAPAPAPSKHKKK 496
Query: 158 KSCPGAATTSEAAQIEQAPAPEVGS 182
GA S + + PAP G+
Sbjct: 497 GKKHGAPAPSPS--LLGPPAPPTGA 565
>TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {Arabidopsis
thaliana;} , partial (22%)
Length = 978
Score = 47.0 bits (110), Expect = 2e-05
Identities = 39/138 (28%), Positives = 56/138 (40%), Gaps = 10/138 (7%)
Frame = +1
Query: 51 PTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKA 110
P+ A AP +SV A + PT + A+SP+S G AV +S T+ +++ A
Sbjct: 46 PSNAQAP-------ASVPANSPPTTLAATSPSS-GTPPAVTQAASPPTTSNPPTISQPPA 201
Query: 111 AASSD----TPIGDKEKENETPKSPPRQDAPPSPPSTHDAGS------MPSPPHQGEKSC 160
+ TP K +PK+P Q PP PP + +P PP
Sbjct: 202 IVAPKPAPVTPPAPKVAPASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLPPPPVATPPPL 381
Query: 161 PGAATTSEAAQIEQAPAP 178
P A+ APAP
Sbjct: 382 PPPKIAPTPAKTSPAPAP 435
>TC218404 UP|Q9MAX7 (Q9MAX7) Epsilon2-COP (Fragment), partial (49%)
Length = 485
Score = 47.0 bits (110), Expect = 2e-05
Identities = 29/93 (31%), Positives = 44/93 (47%)
Frame = +2
Query: 63 AESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKE 122
A + ++ T+P + A+SP S T + A SS AT+ SAS +++ + + P K
Sbjct: 86 ATTCISGRTKPPSTAATSPTSPKKTPSSATPSSTAATSPSASFSSSSPRSMTTLPHLSKP 265
Query: 123 KENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQ 155
+ SPP P PPS +P PP Q
Sbjct: 266 SNSSLSISPP--PTPRIPPSRVSRSGLPIPPLQ 358
>TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light receptor
(Photolyase/blue light photoreceptor PHR2), partial
(84%)
Length = 1641
Score = 46.6 bits (109), Expect = 3e-05
Identities = 41/140 (29%), Positives = 59/140 (41%)
Frame = +3
Query: 45 SNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAS 104
SNP + PT P A AT PT P+++P S+G+ T+AS++
Sbjct: 165 SNPPSPPT----PSTPLSPSLPTAPATLPTPPPSAAPPSSGSAT----------TSASST 302
Query: 105 VNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAA 164
++A+ + TP +P SP PP+ S H A + P+P S P +
Sbjct: 303 MSASPPPTT--TP---------SPSSPSTASTPPTTASRHPASTRPAPTAPPSSSTP--S 443
Query: 165 TTSEAAQIEQAPAPEVGSSS 184
TS AA AP S S
Sbjct: 444 PTSAAASRRAAPISSCASGS 503
Score = 38.1 bits (87), Expect = 0.009
Identities = 18/56 (32%), Positives = 26/56 (46%)
Frame = +3
Query: 123 KENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 178
K P SPP P SP ++P+PP G+ATTS ++ + +P P
Sbjct: 156 KPPSNPPSPPTPSTPLSPSLPTAPATLPTPPPSAAPPSSGSATTSASSTMSASPPP 323
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 46.2 bits (108), Expect = 3e-05
Identities = 42/152 (27%), Positives = 65/152 (42%), Gaps = 1/152 (0%)
Frame = +1
Query: 26 STKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAG 85
S+ P R T +S T P + P+T +P + +S+ A P+A P+++P
Sbjct: 85 SSSPPSPTRTTSPASSQST--PSSPPSTTTSPSPTSPPKST-AKPPSPSA-PSTTPPCRT 252
Query: 86 ATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHD 145
+ + + ++ S S + T A SS + P SPP+ PP+PP
Sbjct: 253 FSRSTPPSTPSRTSSPSTSSSTTSAPRSST-------RSPTAPPSPPQCTKPPAPPPDPP 411
Query: 146 AGSMPSPPHQGEKS-CPGAATTSEAAQIEQAP 176
A S SP EKS P TT+ + Q P
Sbjct: 412 ASS-TSPTSTAEKSHSPPKTTTAHSPQRSSNP 504
>TC217534 similar to GB|AAB62826.1|2252827|IG005I10 coded for by A. thaliana
cDNA N96903 {Arabidopsis thaliana;} , partial (98%)
Length = 1147
Score = 45.8 bits (107), Expect = 4e-05
Identities = 36/130 (27%), Positives = 61/130 (46%), Gaps = 2/130 (1%)
Frame = +3
Query: 51 PTTAAAPK--GKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNAT 108
P+ +++P + + S A + PT AS+ +S+ ATA + + AT+ SA+
Sbjct: 156 PSASSSPS*GSTSTSLSQNALSATPTRCSAST-SSSAATALLRLPPAPPATSTSATTAPL 332
Query: 109 KAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSE 168
+SS P + + T + PPS PS A + P+PP +S AT+S
Sbjct: 333 STPSSSPLPSAARSPASPTASASSPASFPPSLPSPSAATAPPTPPE--SRSSSRGATSSS 506
Query: 169 AAQIEQAPAP 178
A + ++A P
Sbjct: 507 ARRAQRAVNP 536
Score = 30.0 bits (66), Expect = 2.6
Identities = 22/70 (31%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Frame = +3
Query: 24 AESTKAPKRRRLTKASSD--AGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSP 81
A ST A T +SS + +P + +A++P + S +AAT P P S
Sbjct: 300 ATSTSATTAPLSTPSSSPLPSAARSPASPTASASSPASFPPSLPSPSAATAPPTPPESRS 479
Query: 82 ASAGATAAVA 91
+S GAT++ A
Sbjct: 480 SSRGATSSSA 509
>CA853599 weakly similar to PIR|F86379|F86 protein F21J9.28 [imported] -
Arabidopsis thaliana, partial (58%)
Length = 389
Score = 45.8 bits (107), Expect = 4e-05
Identities = 37/111 (33%), Positives = 44/111 (39%), Gaps = 8/111 (7%)
Frame = -1
Query: 82 ASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPP 141
A GA A S T SA + AT AAS+ + TP R PSP
Sbjct: 389 AGDGAGAGQRRRRSWSWTT*SARLTATAMAASASRSSSSSTQRASTPTRSWRTSRTPSPS 210
Query: 142 STHDAGSMPSPPHQGEKSCPGAATTS--------EAAQIEQAPAPEVGSSS 184
ST A + PSPP SC +A T+ AA + A AP SS
Sbjct: 209 STSTA-TAPSPPRSSTPSCAASARTAPSPSAAG*SAASMATATAPSTSRSS 60
>CO985160
Length = 777
Score = 45.8 bits (107), Expect = 4e-05
Identities = 41/117 (35%), Positives = 58/117 (49%), Gaps = 10/117 (8%)
Frame = -1
Query: 10 TEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGT------SNPGAQPTTAAAPK-GKNV 62
T DAK A + A +T A T A+S A T S A T++AP
Sbjct: 735 TSADAKPT--AASSAVATTAAATSHATTATSSAPTTAATTSSAAAAAAATSSAPTTAATT 562
Query: 63 AESSVAAATEPTAVPASSPASAGATAAV---AAESSIGATAASASVNATKAAASSDT 116
+ ++ AAAT +A A++ S+ ATAA AA + ATAA+++ AT AA S+ T
Sbjct: 561 SSAAAAAATTSSAAAAAATTSSAATAAATGPAAAACAAATAAASANPATAAATSTST 391
Score = 43.9 bits (102), Expect = 2e-04
Identities = 36/109 (33%), Positives = 55/109 (50%), Gaps = 7/109 (6%)
Frame = -1
Query: 14 AKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEP 73
A A A ++ AP T +++ A A T++AA + ++ AAAT P
Sbjct: 630 ATTSSAAAAAAATSSAPTTAATTSSAAAA------AATTSSAAAAAATTSSAATAAATGP 469
Query: 74 TAVP-ASSPASAGATAAVAAESSIG------ATAASASVNATKAAASSD 115
A A++ A+A A A AA +S ATAA+++V AT AAA++D
Sbjct: 468 AAAACAAATAAASANPATAAATSTSTTPTTTATAAASAVTATAAAAATD 322
Score = 35.4 bits (80), Expect = 0.061
Identities = 23/49 (46%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Frame = +2
Query: 65 SSVAAATEPTAVPASSPASAG-ATAAVAAESSIGATAASASVNATKAAA 112
++VA E AV A+ A+AG AAVAAE + A AA+ V A AAA
Sbjct: 410 AAVAGFAEAAAVAAAQAAAAGPVAAAVAAEEVVAAAAAAEEVVAAAAAA 556
Score = 35.0 bits (79), Expect = 0.079
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 1/121 (0%)
Frame = -1
Query: 65 SSVAAATEPTAVPASSPASAGATA-AVAAESSIGATAASASVNATKAAASSDTPIGDKEK 123
++ +A +PTA ++ +A AT+ A A SS TAA+ S A AAA+S P
Sbjct: 741 ATTSADAKPTAASSAVATTAAATSHATTATSSAPTTAATTSSAAAAAAATSSAPTTAATT 562
Query: 124 ENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 183
+ + A + +T A + + G + AA T+ AA A A +S
Sbjct: 561 SSAAAAAATTSSAAAAAATTSSAATAAA---TGPAAAACAAATA-AASANPATAAATSTS 394
Query: 184 S 184
+
Sbjct: 393 T 391
Score = 30.4 bits (67), Expect = 2.0
Identities = 30/114 (26%), Positives = 46/114 (40%), Gaps = 5/114 (4%)
Frame = -1
Query: 62 VAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAAS-----ASVNATKAAASSDT 116
+ ES+ A T+ A A++ A A AA +S TA S A+ ++ AAA++ T
Sbjct: 771 IVESAAATNAATTSADAKPTAASSAVATTAAATSHATTATSSAPTTAATTSSAAAAAAAT 592
Query: 117 PIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAA 170
AP + +T A + + + AATTS AA
Sbjct: 591 -----------------SSAPTTAATTSSAAAAAATT---SSAAAAAATTSSAA 490
Score = 29.6 bits (65), Expect = 3.3
Identities = 22/67 (32%), Positives = 30/67 (43%)
Frame = +2
Query: 37 KASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSI 96
+A++ A A P AA AE VAAA V A++ A+ AAV +
Sbjct: 431 EAAAVAAAQAAAAGPVAAAV-----AAEEVVAAAAAAEEVVAAAAAAEEVVAAVVGAEEV 595
Query: 97 GATAASA 103
A AA+A
Sbjct: 596 AAAAAAA 616
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 45.8 bits (107), Expect = 4e-05
Identities = 40/117 (34%), Positives = 45/117 (38%), Gaps = 3/117 (2%)
Frame = +2
Query: 29 APKRRRLTKASSDAGTSNPGAQPTTAAAPKGKN---VAESSVAAATEPTAVPASSPASAG 85
AP + T S S P PTT A P A AA P A P +PA A
Sbjct: 206 APSQPPTTTPSPPPPRSAPAPAPTTPATPPPATPPPAATPPPAATPPPAATP--TPAPAP 379
Query: 86 ATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPS 142
TAA SS A+ S S T + S +TP G P P A P PPS
Sbjct: 380 PTAAPTPASSPAASPPSPSPTVTPSPTSPNTPPG--------PSPGPSGSAEPPPPS 526
Score = 40.4 bits (93), Expect = 0.002
Identities = 34/124 (27%), Positives = 42/124 (33%)
Frame = +2
Query: 47 PGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVN 106
P PTT +P A + A T T PA+ P +A A + T A A
Sbjct: 209 PSQPPTTTPSPPPPRSAPAP-APTTPATPPPATPPPAATPPPAATPPPAATPTPAPAPPT 385
Query: 107 ATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT 166
A ASS TP SP + PP P + P PP + T
Sbjct: 386 AAPTPASSPAASPPSPSPTVTP-SPTSPNTPPGPSPGPSGSAEPPPPSAAFSASKAFIAT 562
Query: 167 SEAA 170
S A
Sbjct: 563 SALA 574
Score = 32.0 bits (71), Expect = 0.67
Identities = 25/96 (26%), Positives = 36/96 (37%)
Frame = +2
Query: 83 SAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPS 142
S T + + GA + ++ + P + T SPP + P+P
Sbjct: 95 STATTTTFSLDMGSGAVQLFLILGLLASSCLAQAPGAAPSQPPTTTPSPPPPRSAPAPAP 274
Query: 143 THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 178
T A P+ P + P AAT AA APAP
Sbjct: 275 TTPATPPPATPPPA-ATPPPAATPPPAATPTPAPAP 379
>TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_40, partial
(76%)
Length = 1447
Score = 45.4 bits (106), Expect = 6e-05
Identities = 44/148 (29%), Positives = 70/148 (46%), Gaps = 6/148 (4%)
Frame = +1
Query: 50 QPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGA----TAAVAAESSIGAT--AASA 103
Q T P A ++ + AT P+ + A+S ASA T++ + S+ T A+SA
Sbjct: 313 QETIQDVPHPPRHAYTASSTATVPSQL-ATSAASASP*PPPTSSTTSSSAAARTTRASSA 489
Query: 104 SVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGA 163
+ A+ A+A++ +P +K N SP PP+PP+T S +PP P A
Sbjct: 490 APPASLASAATPSPSSNKLPPNTVKSSPTASPPPPAPPAT----SPSAPP-------PQA 636
Query: 164 ATTSEAAQIEQAPAPEVGSSSYYNMLPN 191
AT+S P+P S+ +LP+
Sbjct: 637 ATSSTP---PSPPSPAAPPSTVSILLPS 711
Score = 34.7 bits (78), Expect = 0.10
Identities = 44/154 (28%), Positives = 62/154 (39%), Gaps = 10/154 (6%)
Frame = +1
Query: 52 TTAAAPKGKNVAESSVAAATEPTAVPASSPASAGAT-AAVAAESSIGATAASASVNATK- 109
+TA P + +S + PT+ SS A+A T A+ AA + A+AA+ S ++ K
Sbjct: 367 STATVPSQLATSAASASP*PPPTSSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKL 546
Query: 110 ---AAASSDTPIGDKEKENET-PKSPPRQDA----PPSPPSTHDAGSMPSPPHQGEKSCP 161
SS T T P +PP Q A PPSPPS +PP P
Sbjct: 547 PPNTVKSSPTASPPPPAPPATSPSAPPPQAATSSTPPSPPSP------AAPPSTVSILLP 708
Query: 162 GAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEP 195
S + P V SS+ P + +P
Sbjct: 709 SP*AVSNSQSPPPPSPPAVQSSTLAP*SPASPQP 810
Score = 32.7 bits (73), Expect = 0.39
Identities = 35/150 (23%), Positives = 57/150 (37%), Gaps = 21/150 (14%)
Frame = +1
Query: 24 AESTKAPKRRRLTKASSDAGTSN--PGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSP 81
+ +T + R T+ASS A ++ A P+ ++ N +SS A+ P A PA+SP
Sbjct: 436 SSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKLPPNTVKSSPTASPPPPAPPATSP 615
Query: 82 -------------------ASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKE 122
A+ +T ++ S + + + + A S T
Sbjct: 616 SAPPPQAATSSTPPSPPSPAAPPSTVSILLPSP*AVSNSQSPPPPSPPAVQSST------ 777
Query: 123 KENETPKSPPRQDAPPSPPSTHDAGSMPSP 152
+P SP AP PPS + P P
Sbjct: 778 LAP*SPASPQPPTAPSDPPSVKACQNTPPP 867
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 45.4 bits (106), Expect = 6e-05
Identities = 34/138 (24%), Positives = 51/138 (36%), Gaps = 3/138 (2%)
Frame = +2
Query: 30 PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKN---VAESSVAAATEPTAVPASSPASAGA 86
P T S + TS+P +T +APK A S + P A P + PA++ +
Sbjct: 215 PPSSTSTSPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTS 394
Query: 87 TAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDA 146
+ A S+ +A + + S P ++ P +PP PP PP
Sbjct: 395 PTSRPARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKSAPPLAPPTPRHPPPPP----- 559
Query: 147 GSMPSPPHQGEKSCPGAA 164
PP CP A
Sbjct: 560 -----PPST*SPLCPNKA 598
Score = 43.9 bits (102), Expect = 2e-04
Identities = 42/158 (26%), Positives = 67/158 (41%), Gaps = 23/158 (14%)
Frame = +2
Query: 44 TSNPGAQPTTAAAPKGKNVAES------SVAAATEPT---AVPASSPASAGATAAVAAES 94
T++P P T A+P + S S AA P+ +P P+S + + +++
Sbjct: 80 TTSPACWPHTPASPPSTTTSPSPTWPKRSTAARPSPSWPSTMPPCPPSSTSTSPSPPSKT 259
Query: 95 SIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPP--------------SP 140
S +T++S + +A K++ S T + P PP APP SP
Sbjct: 260 SSPSTSSSTT-SAPKSSTRSTTAPPSSPPCSRPPAPPP---APPATSTSPTSRPARSASP 427
Query: 141 PSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 178
P T A S PS + K CP + +S++A P P
Sbjct: 428 PKTTTAPSTPSTSNPSRK-CPTTSPSSKSAPPLAPPTP 538
>TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-phosphate
isomerase (Phosphoriboisomerase) , partial (47%)
Length = 729
Score = 45.4 bits (106), Expect = 6e-05
Identities = 40/150 (26%), Positives = 66/150 (43%), Gaps = 16/150 (10%)
Frame = +2
Query: 51 PTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATA-----ASASV 105
PT ++P K + A++T P P+SSP + + SS + A A A +
Sbjct: 278 PTPISSPPRK--PPWTPASSTPPPPPPSSSPKTTSRKSPPTRPSSTSSPAWSSASAPAPL 451
Query: 106 NATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCP---- 161
+T + AS+ + + K + PP+ PSP ++ S P+PP + P
Sbjct: 452 PSTPSTASASSSAKENSKTSSESPPPPKPTTRPSPSASPSPISTPTPPSISPSTAPTRSI 631
Query: 162 GAATTSEAA-------QIEQAPAPEVGSSS 184
++T+S AA + +APA SSS
Sbjct: 632 PSSTSSRAAAAPSFEKRWSRAPARSSSSSS 721
>TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (47%)
Length = 870
Score = 45.1 bits (105), Expect = 8e-05
Identities = 39/145 (26%), Positives = 57/145 (38%)
Frame = +3
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIG 97
A S+ + A P A + K+ A + ++ P A PAS+PA++ T A
Sbjct: 150 APSNTPATPAAATPAQAPSTVPKSPAPVASPKSSPPAATPASTPAASPTTTPAAPAPVTK 329
Query: 98 ATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGE 157
AAS A P+ P S P P + P+T A S P+P H+ +
Sbjct: 330 PPAASPPPATPPPATPPPAPVPVSSPPAPVPVSSPPAPVPVAAPTTPVAPS-PAPKHKKK 506
Query: 158 KSCPGAATTSEAAQIEQAPAPEVGS 182
GA S + PAP G+
Sbjct: 507 GKKHGAPAPS---PLLGPPAPPTGA 572
Score = 43.5 bits (101), Expect = 2e-04
Identities = 44/141 (31%), Positives = 60/141 (42%), Gaps = 4/141 (2%)
Frame = +3
Query: 43 GTSNPGAQPT-TAAAPKGKNVAE--SSVAAATEPTAVPASSPASAGATAAVAAESSIGAT 99
G +P A P+ T A P A+ S+V + P A P SSP +A + AA + T
Sbjct: 129 GGQSPAAAPSNTPATPAAATPAQAPSTVPKSPAPVASPKSSPPAATPASTPAASPT---T 299
Query: 100 AASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAP-PSPPSTHDAGSMPSPPHQGEK 158
+A TK A+S P P +PP P SPP+ S P+P
Sbjct: 300 TPAAPAPVTKPPAASPPPA------TPPPATPPPAPVPVSSPPAPVPVSSPPAP------ 443
Query: 159 SCPGAATTSEAAQIEQAPAPE 179
P AA T+ A +PAP+
Sbjct: 444 -VPVAAPTTPVA---PSPAPK 494
Score = 43.1 bits (100), Expect = 3e-04
Identities = 49/152 (32%), Positives = 62/152 (40%), Gaps = 7/152 (4%)
Frame = +3
Query: 23 QAEST--KAPKRRRLTKASSDAGT--SNPGAQPTT---AAAPKGKNVAESSVAAATEPTA 75
QA ST K+P K+S A T S P A PTT A AP K A +S AT P A
Sbjct: 195 QAPSTVPKSPAPVASPKSSPPAATPASTPAASPTTTPAAPAPVTKPPA-ASPPPATPPPA 371
Query: 76 VPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQD 135
P +P + A SS A A+ T A S P K+ + +P
Sbjct: 372 TPPPAPVPVSSPPAPVPVSSPPAPVPVAA--PTTPVAPSPAPKHKKKGKKHGAPAPSPLL 545
Query: 136 APPSPPSTHDAGSMPSPPHQGEKSCPGAATTS 167
PP+PP+ P P + S PG T +
Sbjct: 546 GPPAPPT-----GAPGPSE--DASSPGPGTAA 620
>TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, partial (46%)
Length = 829
Score = 44.7 bits (104), Expect = 1e-04
Identities = 41/152 (26%), Positives = 60/152 (38%), Gaps = 12/152 (7%)
Frame = +1
Query: 45 SNPGAQPTTAAAPKGKNV------AESSVAAATEPTAVPASSPASAGATAAVAAESSIGA 98
SNP P T + + + + SV + +P P+S + S
Sbjct: 118 SNPTQPPNTKTFLQNQAIHSQ**CLKPSVPMSHPKPPIPTPPPSSTSPSIRTPGASPPPP 297
Query: 99 TAASASVNATKAAASS---DTPIGDKEK---ENETPKSPPRQDAPPSPPSTHDAGSMPSP 152
TAASAS AT +A SS P + SP APP P+T +P+
Sbjct: 298 TAASASTTATPSARSSAATSAPAAASASSTCSSAATSSPSSAAAPPPTPAT-----LPTR 462
Query: 153 PHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 184
G +CP A+ +S +A+ +A A SS
Sbjct: 463 S*SGTTTCPAASASSPSARRSRASASAATESS 558
Score = 32.3 bits (72), Expect = 0.51
Identities = 37/135 (27%), Positives = 55/135 (40%), Gaps = 22/135 (16%)
Frame = +1
Query: 38 ASSDAGTSNPGAQP---TTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAES 94
+S+ PGA P T A+A + S AA + P A ASS S+ AT++ ++ +
Sbjct: 247 SSTSPSIRTPGASPPPPTAASASTTATPSARSSAATSAPAAASASSTCSSAATSSPSSAA 426
Query: 95 SIGAT----------------AASAS---VNATKAAASSDTPIGDKEKENETPKSPPRQD 135
+ T AASAS ++A+AS+ T + SP R
Sbjct: 427 APPPTPATLPTRS*SGTTTCPAASASSPSARRSRASASAAT--------ESSSSSPTRSS 582
Query: 136 APPSPPSTHDAGSMP 150
+ SP S S P
Sbjct: 583 STTSPISKCCTKSRP 627
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 44.3 bits (103), Expect = 1e-04
Identities = 30/79 (37%), Positives = 45/79 (55%), Gaps = 1/79 (1%)
Frame = +1
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVA-AATEPTAVPASSPASAGATAAVAAESSI 96
A+S A T+ A AA+ A SS AAT P A +++PA+A A A+ + +
Sbjct: 79 ATSHAATTTSSAAAAAAASSAATAAATSSATTAATAPAAAASATPATA-AAASTSTTPTT 255
Query: 97 GATAASASVNATKAAASSD 115
ATAA+++V AT A A++D
Sbjct: 256 TATAAASAVAATAATAATD 312
Score = 43.9 bits (102), Expect = 2e-04
Identities = 27/75 (36%), Positives = 44/75 (58%)
Frame = +1
Query: 40 SDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGAT 99
S A T+ A+PT ++ A +S AA T +A A++ AS+ ATAA + ++ AT
Sbjct: 7 SAAATAPADAKPTATSSAVATTAAATSHAATTTSSAAAAAA-ASSAATAAATSSATTAAT 183
Query: 100 AASASVNATKAAASS 114
A +A+ +AT A A++
Sbjct: 184 APAAAASATPATAAA 228
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/74 (39%), Positives = 38/74 (51%)
Frame = +1
Query: 44 TSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASA 103
T+ A TTAAA SS AA A ASS A+A AT++ ++ A AASA
Sbjct: 43 TATSSAVATTAAATSHAATTTSSAAA-----AAAASSAATAAATSSATTAATAPAAAASA 207
Query: 104 SVNATKAAASSDTP 117
+ AA++S TP
Sbjct: 208 TPATAAAASTSTTP 249
Score = 33.9 bits (76), Expect = 0.18
Identities = 27/103 (26%), Positives = 41/103 (39%)
Frame = +1
Query: 68 AAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENET 127
AAAT P ++ +SA AT A A AA+ + +A AAA+S T
Sbjct: 10 AAATAPADAKPTATSSAVATTAAATSH-----AATTTSSAAAAAAASSAATAAATSSATT 174
Query: 128 PKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAA 170
+ P A +P + A + +P + A T+ A
Sbjct: 175 AATAPAAAASATPATAAAASTSTTPTTTATAAASAVAATAATA 303
Score = 33.1 bits (74), Expect = 0.30
Identities = 25/70 (35%), Positives = 34/70 (47%), Gaps = 5/70 (7%)
Frame = +1
Query: 38 ASSDAGTSNPGAQPTTAA-APKGKNVAESSVAAA----TEPTAVPASSPASAGATAAVAA 92
A+S A T+ + TTAA AP A + AAA T PT ++ ++ ATAA AA
Sbjct: 127 AASSAATAAATSSATTAATAPAAAASATPATAAAASTSTTPTTTATAAASAVAATAATAA 306
Query: 93 ESSIGATAAS 102
G +S
Sbjct: 307 TDGWGRNGSS 336
Score = 28.9 bits (63), Expect = 5.7
Identities = 22/68 (32%), Positives = 27/68 (39%), Gaps = 4/68 (5%)
Frame = -2
Query: 47 PGAQPTTAAAPKGKNVAESSVAAATEPTAVPA----SSPASAGATAAVAAESSIGATAAS 102
P A A + A + V E A A + A+AGA AAV AE A AA
Sbjct: 314 PSVAAVAAVAATAEAAAVAVVVGVVEVEAAAAVAGVAEAAAAGAVAAVVAEEVAAAVAAE 135
Query: 103 ASVNATKA 110
+ A A
Sbjct: 134 EAAAAAAA 111
>BI969707
Length = 711
Score = 43.5 bits (101), Expect = 2e-04
Identities = 45/157 (28%), Positives = 66/157 (41%), Gaps = 6/157 (3%)
Frame = +3
Query: 17 RGGAENQAESTKAPKR--RRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAA-ATEP 73
RG E ++ T R R+TK ++ + A T A N + S+ + A+
Sbjct: 174 RGDCEGESAKTA*YARFWGRITKGTTWSSPQKS*AHRCTFVARAAHNCSASATQSWASHC 353
Query: 74 TAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGD-KEKENETPKSPP 132
+A P P G G T+++AS + +A TP E TP SPP
Sbjct: 354 SASPPPVPCDRGT----------GNTSSTAS--SPRAPPPRSTPSEPCSEPPPHTPPSPP 497
Query: 133 R--QDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTS 167
R + APPS + S P PP +G S P A++S
Sbjct: 498 RIYRSAPPSXMTPPPQSSTPXPPPRGPNSPPSTASSS 608
Score = 32.3 bits (72), Expect = 0.51
Identities = 28/124 (22%), Positives = 45/124 (35%), Gaps = 2/124 (1%)
Frame = +3
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIG 97
+++ + S+ A P +G S+ ++ P P S+P+ + S
Sbjct: 324 SATQSWASHCSASPPPVPCDRGTGNTSSTASSPRAPP--PRSTPSEPCSEPPPHTPPSPP 497
Query: 98 ATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAP--PSPPSTHDAGSMPSPPHQ 155
SA + S TP N P + +P SPPST A SPP
Sbjct: 498 RIYRSAPPSXMTPPPQSSTPXPPPRGPNSPPSTASSSPSPLRGSPPSTCSAXXPKSPPRS 677
Query: 156 GEKS 159
E++
Sbjct: 678 PERA 689
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.304 0.121 0.318
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,374,470
Number of Sequences: 63676
Number of extensions: 247959
Number of successful extensions: 6329
Number of sequences better than 10.0: 707
Number of HSP's better than 10.0 without gapping: 3620
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5179
length of query: 507
length of database: 12,639,632
effective HSP length: 101
effective length of query: 406
effective length of database: 6,208,356
effective search space: 2520592536
effective search space used: 2520592536
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0201.4


