

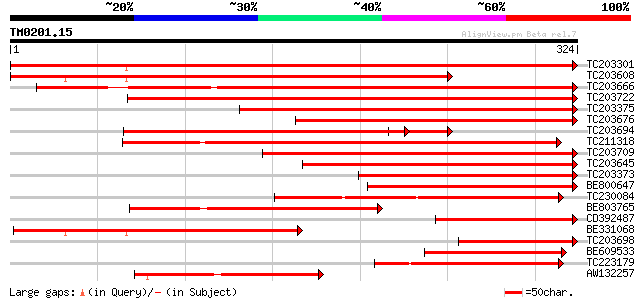
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.15
(324 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203301 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, com... 555 e-158
TC203608 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, parti... 402 e-112
TC203666 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , p... 361 e-100
TC203722 similar to UP|CAHC_ARATH (P27140) Carbonic anhydrase, c... 356 9e-99
TC203375 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 352 1e-97
TC203676 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 295 2e-80
TC203694 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, parti... 278 2e-75
TC211318 UP|Q9XG11 (Q9XG11) Carbonic anhydrase , partial (95%) 278 2e-75
TC203709 similar to UP|CAH2_FLALI (P46513) Carbonic anhydrase 2 ... 255 2e-68
TC203645 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , p... 243 9e-65
TC203373 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 233 9e-62
BE800647 similar to GP|8954289|gb| carbonic anhydrase {Vigna rad... 189 1e-48
TC230084 weakly similar to GB|AAM65957.1|21594039|AY088420 carbo... 159 2e-39
BE803765 weakly similar to GP|27652184|gb carbonic anhydrase 2 {... 154 5e-38
CD392487 153 1e-37
BE331068 weakly similar to GP|8954289|gb| carbonic anhydrase {Vi... 139 1e-33
TC203698 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 129 2e-30
BE609533 similar to GP|8954289|gb|A carbonic anhydrase {Vigna ra... 113 1e-25
TC223179 weakly similar to UP|O81875 (O81875) Carbonate dehydrat... 108 3e-24
AW132257 similar to GP|8569250|pdb| Chain A The X-Ray Crystallo... 108 3e-24
>TC203301 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, complete
Length = 1413
Score = 555 bits (1430), Expect = e-158
Identities = 280/328 (85%), Positives = 307/328 (93%), Gaps = 4/328 (1%)
Frame = +1
Query: 1 MSTSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSS-SSSSSFPSLIQDRPVFAAPA 59
MSTS+ING+ LSSISPAKTSL++ TLRP V A+ ++PSS SSSSSFPSLIQDRPVFAAPA
Sbjct: 52 MSTSSINGWCLSSISPAKTSLRKATLRPSVFATLNTPSSPSSSSSFPSLIQDRPVFAAPA 231
Query: 60 PII-PTL--DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEA 116
PII PT+ DM K YE+AIEELQKLLREK+ELKATAAEKVEQITA LGT+SSDGIPSSEA
Sbjct: 232 PIITPTVREDMAKEYEKAIEELQKLLREKSELKATAAEKVEQITASLGTSSSDGIPSSEA 411
Query: 117 SDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVV 176
SDRIK GF++FKKEKYDKNPALYGELAKGQSP +MVFACSDSRVCPSHVLDFQPGEAFVV
Sbjct: 412 SDRIKAGFIHFKKEKYDKNPALYGELAKGQSPKFMVFACSDSRVCPSHVLDFQPGEAFVV 591
Query: 177 RNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTD 236
RNVAN+VPPYDQ+K++G+GAA+EYAVLHLKVS IVVIGHSACGGIKGLLSFP+DGT STD
Sbjct: 592 RNVANIVPPYDQSKYAGTGAAVEYAVLHLKVSEIVVIGHSACGGIKGLLSFPYDGTYSTD 771
Query: 237 FIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLA 296
FIE+WV IGLPAKAKVK +HGDAPF ELC+HCEKEAVNVSLGNLL+YPFVR+GLVNKTL+
Sbjct: 772 FIEEWVKIGLPAKAKVKTQHGDAPFAELCSHCEKEAVNVSLGNLLTYPFVRDGLVNKTLS 951
Query: 297 LKGGYYDFVKGSFELWSLQFDLASSFSV 324
LKGGYYDFVKGSFELW LQF LASSFSV
Sbjct: 952 LKGGYYDFVKGSFELWGLQFGLASSFSV 1035
>TC203608 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (78%)
Length = 812
Score = 402 bits (1032), Expect = e-112
Identities = 209/259 (80%), Positives = 232/259 (88%), Gaps = 6/259 (2%)
Frame = +3
Query: 1 MSTSTINGFYLSSISPAKTSLKRVTLRPLV---VASASSPSSSSSSSFPSLIQDRPVFAA 57
MSTS+ING+ LSSISPAKTSLK+ TL P V V++ SSPSSSSSSSFPSLIQDRPVFAA
Sbjct: 36 MSTSSINGWCLSSISPAKTSLKKATLGPSVFATVSTPSSPSSSSSSSFPSLIQDRPVFAA 215
Query: 58 PAPII-PTL--DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSS 114
PAPII PT+ DM K YE+AIEELQKLLREK+ELKATAAEKVEQITA LGT+SSDGIPSS
Sbjct: 216 PAPIITPTVREDMAKEYEQAIEELQKLLREKSELKATAAEKVEQITASLGTSSSDGIPSS 395
Query: 115 EASDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAF 174
EASDRIK GF++FKKEKYDKNPALYGELAKGQSP +MVFACSDSRVCPSHVLDFQPGEAF
Sbjct: 396 EASDRIKAGFIHFKKEKYDKNPALYGELAKGQSPKFMVFACSDSRVCPSHVLDFQPGEAF 575
Query: 175 VVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTS 234
VVRNVAN+ PPYDQ+K++G+GAA+EYAV HLK S+I IGH ACGGIKGLLS P+DGT S
Sbjct: 576 VVRNVANIDPPYDQSKYAGTGAALEYAVSHLKESDIEDIGHRACGGIKGLLSLPYDGTYS 755
Query: 235 TDFIEDWVSIGLPAKAKVK 253
TD I++W +IGLPAKA VK
Sbjct: 756 TDLIDEWRTIGLPAKANVK 812
>TC203666 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , partial (80%)
Length = 1194
Score = 361 bits (927), Expect = e-100
Identities = 186/309 (60%), Positives = 222/309 (71%)
Frame = +2
Query: 16 PAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKNYEEAI 75
P T+ K + + +S SS S + L Q + A G++YEEAI
Sbjct: 41 PLYTANKNSNSHSRITNTHTSLFSSKSHQYLYLAQKKEEMA-----------GESYEEAI 187
Query: 76 EELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFKKEKYDKN 135
L KLL EK +L AA K++ +TA+L S E RI+TGF+ FK EK++KN
Sbjct: 188 ASLTKLLSEKADLGGVAAAKIKDLTAELDAAGSKPFNPEE---RIRTGFIQFKNEKFEKN 358
Query: 136 PALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSG 195
P LYGELAKGQSP +MVFACSDSRVCPSH+LDFQPGEAF+VRN+ANMVPPYD+ K+SG+G
Sbjct: 359 PDLYGELAKGQSPKFMVFACSDSRVCPSHILDFQPGEAFMVRNIANMVPPYDKTKYSGAG 538
Query: 196 AAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAK 255
AAIEYAVLHLKV NIVVIGHS CGGIKGL+S P DGTT+++FIE WV I PAK+KVKA
Sbjct: 539 AAIEYAVLHLKVENIVVIGHSCCGGIKGLMSIPDDGTTASEFIEQWVQICTPAKSKVKAG 718
Query: 256 HGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQ 315
D F E CT+CEKEAVNVSLGNLL+YPFVR+G+VNKTLALKG +YDFV G+FELW L
Sbjct: 719 KSDLSFSEQCTNCEKEAVNVSLGNLLTYPFVRDGVVNKTLALKGAHYDFVNGNFELWDLN 898
Query: 316 FDLASSFSV 324
F L + SV
Sbjct: 899 FKLLPTISV 925
>TC203722 similar to UP|CAHC_ARATH (P27140) Carbonic anhydrase, chloroplast
precursor (Carbonate dehydratase) , partial (69%)
Length = 1120
Score = 356 bits (913), Expect = 9e-99
Identities = 172/258 (66%), Positives = 208/258 (79%), Gaps = 1/258 (0%)
Frame = +1
Query: 68 GKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYF 127
G +YEEAI L KL+ EK +L AA K++Q+TA+L T +++G +RI+TGF +F
Sbjct: 142 GGSYEEAIAALTKLISEKADLSGVAAAKIKQLTAELETATANGSTPFNPDERIRTGFAHF 321
Query: 128 KKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYD 187
K EKY KNP LYGELAKGQSP +MVFACSDSRVCPSH+LDF PGEAFVVRN+ANMVPPYD
Sbjct: 322 KNEKYQKNPELYGELAKGQSPKFMVFACSDSRVCPSHILDFNPGEAFVVRNIANMVPPYD 501
Query: 188 QAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLP 247
+ K+SG+GAAIEYAV+HLKV NIVVIGHS CGGIKGL+S P DGTT+++FIE WV I P
Sbjct: 502 KTKYSGTGAAIEYAVVHLKVENIVVIGHSCCGGIKGLMSIPDDGTTASEFIEHWVQICTP 681
Query: 248 AKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLV-NKTLALKGGYYDFVK 306
AK+KVK + F E CT+CEKEAVNVSLGNLL+YPFVR+ +V KTLALKG +Y+FVK
Sbjct: 682 AKSKVKTEASTLEFSEQCTNCEKEAVNVSLGNLLTYPFVRDAVVKKKTLALKGAHYNFVK 861
Query: 307 GSFELWSLQFDLASSFSV 324
G+FELW L F +++S SV
Sbjct: 862 GTFELWDLDFKISNSVSV 915
>TC203375 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (57%)
Length = 1427
Score = 352 bits (903), Expect = 1e-97
Identities = 166/193 (86%), Positives = 183/193 (94%)
Frame = +3
Query: 132 YDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKF 191
+ ++ ALYGELAKGQSP +MVFACSDSRVCPSHVLDFQPGEAFVVRNVAN+VPPYDQ+K+
Sbjct: 333 FQRHEALYGELAKGQSPKFMVFACSDSRVCPSHVLDFQPGEAFVVRNVANIVPPYDQSKY 512
Query: 192 SGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAK 251
+G+GAA+EYAVLHLKVS IVVIGHSACGGIKGLLSFP+DGT STDFIE+WV IGLPAKAK
Sbjct: 513 AGTGAAVEYAVLHLKVSEIVVIGHSACGGIKGLLSFPYDGTYSTDFIEEWVKIGLPAKAK 692
Query: 252 VKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFEL 311
VK +HGDAPF ELC+HCEKEAVNVSLGNLL+YPFVR+GLVNKTL+LKGGYYDFVKGSFEL
Sbjct: 693 VKTQHGDAPFAELCSHCEKEAVNVSLGNLLTYPFVRDGLVNKTLSLKGGYYDFVKGSFEL 872
Query: 312 WSLQFDLASSFSV 324
W LQF LASSFSV
Sbjct: 873 WGLQFGLASSFSV 911
>TC203676 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (49%)
Length = 790
Score = 295 bits (755), Expect = 2e-80
Identities = 140/161 (86%), Positives = 153/161 (94%)
Frame = +3
Query: 164 HVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKG 223
HVLDFQPGEAFVVRNVAN+VPPYDQ+K++G+GAA+EYAVLHLKVS IVVIGHSACG IKG
Sbjct: 3 HVLDFQPGEAFVVRNVANIVPPYDQSKYAGTGAAVEYAVLHLKVSEIVVIGHSACGVIKG 182
Query: 224 LLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSY 283
LLSFP+DGT STDFIE+WV IGLPAKAKVK +HGDAPF ELC+HCEKEAVNVSLGNLL+Y
Sbjct: 183 LLSFPYDGTYSTDFIEEWVKIGLPAKAKVKTQHGDAPFAELCSHCEKEAVNVSLGNLLTY 362
Query: 284 PFVREGLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
PFVR+GLVNKTL+LKGGYYDFVKGSFELW LQF LASSFSV
Sbjct: 363 PFVRDGLVNKTLSLKGGYYDFVKGSFELWGLQFGLASSFSV 485
>TC203694 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (58%)
Length = 731
Score = 278 bits (712), Expect = 2e-75
Identities = 136/163 (83%), Positives = 149/163 (90%)
Frame = +3
Query: 66 DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFL 125
DM K YE+AIEELQKLLREK+ELKATAAEKVEQITA LGT+SSDGIPSSEASDRIK GF+
Sbjct: 111 DMAKEYEQAIEELQKLLREKSELKATAAEKVEQITASLGTSSSDGIPSSEASDRIKAGFI 290
Query: 126 YFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPP 185
+FKKEKYDKNPALYGELAKGQSP +MVFACSDSRVCPSHVLDFQPGEAFVVRNVAN+VPP
Sbjct: 291 HFKKEKYDKNPALYGELAKGQSPKFMVFACSDSRVCPSHVLDFQPGEAFVVRNVANIVPP 470
Query: 186 YDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFP 228
YDQ+K++G+GAA+EYAVLHL+VS IVVIGH G KG FP
Sbjct: 471 YDQSKYAGTGAAVEYAVLHLRVSEIVVIGHMGLWGYKGARGFP 599
Score = 53.9 bits (128), Expect = 1e-07
Identities = 25/37 (67%), Positives = 27/37 (72%)
Frame = +1
Query: 217 ACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVK 253
ACGGIKGL +FP DG STD IE+WV G P KA VK
Sbjct: 565 ACGGIKGLGAFPCDGAGSTDXIEEWVKSGWPCKAXVK 675
>TC211318 UP|Q9XG11 (Q9XG11) Carbonic anhydrase , partial (95%)
Length = 846
Score = 278 bits (712), Expect = 2e-75
Identities = 141/251 (56%), Positives = 178/251 (70%)
Frame = +3
Query: 65 LDMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGF 124
+ M K+ E AIEEL+KLLR+K E A AA KVE++ A+L P A RI GF
Sbjct: 93 ITMAKSSELAIEELKKLLRDKEEFDAVAAAKVEELIAELQGCGPR--PYEPAVQRIVDGF 266
Query: 125 LYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVP 184
+FK +DKN LY +LA GQ P Y+VF+CSDSRV + +L+FQPGEAF+VRN+ANMVP
Sbjct: 267 THFKINNFDKNSDLYSQLANGQRPKYLVFSCSDSRVSATTILNFQPGEAFMVRNIANMVP 446
Query: 185 PYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSI 244
P++Q ++SG GAAIEYA+ LKV NI+VIGHS CGGI+ L+S P DG+ DFI+DWV I
Sbjct: 447 PFNQLRYSGVGAAIEYAITALKVPNILVIGHSRCGGIQRLMSHPEDGSAPFDFIDDWVKI 626
Query: 245 GLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDF 304
GLPAK KV ++ F E C CEKE+VN SL NL +YP+V +G+ NK +AL GGYYDF
Sbjct: 627 GLPAKLKVLKEYESYDFKEQCKFCEKESVNNSLVNLKTYPYVEKGIRNKNIALLGGYYDF 806
Query: 305 VKGSFELWSLQ 315
V G F+LW +
Sbjct: 807 VIGEFKLWKYE 839
>TC203709 similar to UP|CAH2_FLALI (P46513) Carbonic anhydrase 2 (Carbonate
dehydratase 2) (Fragment) , partial (93%)
Length = 929
Score = 255 bits (651), Expect = 2e-68
Identities = 124/180 (68%), Positives = 144/180 (79%)
Frame = +3
Query: 145 GQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLH 204
GQS V ACSDSR PS +LDF PGEAFVVRN+ANMVPPYD+ K+SG+GAAIEYAVLH
Sbjct: 3 GQSXKXXVXACSDSRXXPSXILDFNPGEAFVVRNIANMVPPYDKTKYSGTGAAIEYAVLH 182
Query: 205 LKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGEL 264
LKV NIVVIGHS CGGIKGL+S P DGTT+++FIE WV I PAK+KVK + F E
Sbjct: 183 LKVENIVVIGHSCCGGIKGLMSIPDDGTTASEFIEHWVQICTPAKSKVKTEANTLEFSEQ 362
Query: 265 CTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
CT CEKEAVNVSLGNLL+Y FVR+ +V KTLALKG +Y+FVKG+FELW L +++S SV
Sbjct: 363 CTSCEKEAVNVSLGNLLTYRFVRDAVVKKTLALKGAHYNFVKGTFELWDLDLKISNSVSV 542
>TC203645 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , partial (53%)
Length = 696
Score = 243 bits (620), Expect = 9e-65
Identities = 116/157 (73%), Positives = 133/157 (83%)
Frame = +1
Query: 168 FQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSF 227
FQPGEAF+VRN+ANMVPPYD+ K+SG+GAAIEYAVLHLKV NIVVIGHS CGGIKGL+S
Sbjct: 1 FQPGEAFMVRNIANMVPPYDKTKYSGAGAAIEYAVLHLKVENIVVIGHSCCGGIKGLMSI 180
Query: 228 PFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVR 287
P DGTT+++FIE WV I PAK+KVKA D F E CT+CEKEAVNVSLGNLL+YPFVR
Sbjct: 181 PDDGTTASEFIEQWVQICTPAKSKVKAGPSDLSFSEQCTNCEKEAVNVSLGNLLTYPFVR 360
Query: 288 EGLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
+G+VNKTLALKG +YDFV G+FELW L F L + SV
Sbjct: 361 DGVVNKTLALKGAHYDFVNGTFELWDLNFKLFPTVSV 471
>TC203373 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (38%)
Length = 717
Score = 233 bits (594), Expect = 9e-62
Identities = 111/125 (88%), Positives = 118/125 (93%)
Frame = +3
Query: 200 YAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDA 259
YAVLHLKVS IVVIGHSACGGIKGLLSFP+DGT STDFIE+WV IGLPAKAKVK +HGDA
Sbjct: 3 YAVLHLKVSEIVVIGHSACGGIKGLLSFPYDGTYSTDFIEEWVKIGLPAKAKVKTQHGDA 182
Query: 260 PFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQFDLA 319
PF ELC+HCEKEAVNVSLGNLL+YPFVR+GLVNKTL+LKGGYYDFVKGSFELW LQF LA
Sbjct: 183 PFAELCSHCEKEAVNVSLGNLLTYPFVRDGLVNKTLSLKGGYYDFVKGSFELWGLQFGLA 362
Query: 320 SSFSV 324
SSFSV
Sbjct: 363 SSFSV 377
>BE800647 similar to GP|8954289|gb| carbonic anhydrase {Vigna radiata},
partial (36%)
Length = 361
Score = 189 bits (481), Expect = 1e-48
Identities = 91/120 (75%), Positives = 106/120 (87%)
Frame = +2
Query: 205 LKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGEL 264
LKVS IVVIGHSACGGIKGLLSFP++GT STDFIE+WV IGLPAKAKVK +H D PF +L
Sbjct: 2 LKVSEIVVIGHSACGGIKGLLSFPYNGTYSTDFIEEWVKIGLPAKAKVKTQHRDTPFTKL 181
Query: 265 CTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
+HC+KEAVNVSLGNLL+YPF+R+ L+NKTL+LKG YY+F+K SF L SLQF+L SSFSV
Sbjct: 182 YSHCKKEAVNVSLGNLLTYPFIRDSLINKTLSLKGRYYNFIKESFNL*SLQFNLTSSFSV 361
>TC230084 weakly similar to GB|AAM65957.1|21594039|AY088420 carbonate
dehydratase-like protein {Arabidopsis thaliana;} ,
partial (54%)
Length = 831
Score = 159 bits (401), Expect = 2e-39
Identities = 80/165 (48%), Positives = 111/165 (66%)
Frame = +3
Query: 152 VFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIV 211
V AC+DSRVCPS++L FQPGE F++RN+AN+VP S AA+++AV L+V NI+
Sbjct: 6 VIACADSRVCPSNILGFQPGEVFMIRNIANLVPVMKNGP-SECNAALQFAVTTLQVENIL 182
Query: 212 VIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKE 271
VIGHS+C GI+ L++ D S +FI WV+ G AK + KA F + C CEKE
Sbjct: 183 VIGHSSCAGIEALMNMQEDAE-SRNFIHKWVANGKLAKQRTKAATAHLSFDQQCKFCEKE 359
Query: 272 AVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQF 316
++N SL NLLSYP++++ + + L+L GGYY+F SFE W+L F
Sbjct: 360 SINQSLLNLLSYPWIQDRVRKELLSLHGGYYNFSNCSFEKWTLDF 494
>BE803765 weakly similar to GP|27652184|gb carbonic anhydrase 2 {Flaveria
bidentis}, partial (43%)
Length = 442
Score = 154 bits (389), Expect = 5e-38
Identities = 72/145 (49%), Positives = 105/145 (71%)
Frame = +2
Query: 69 KNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFK 128
++Y+E+I L KLL EK +L A K++++TA+L TT S + I+T F++FK
Sbjct: 17 ESYQESITSLTKLLSEKAKLNGITAAKIKELTAELDTTGSK---PGNPEEMIRTRFIHFK 187
Query: 129 KEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQ 188
EK+ KNP LYG+LAK QSP ++VF CSDSR+ PSH+L+FQP +AF+VRN+ NM+ PY +
Sbjct: 188 NEKFKKNPYLYGKLAKSQSPKFIVFTCSDSRIYPSHILNFQPSKAFIVRNITNMIAPYYK 367
Query: 189 AKFSGSGAAIEYAVLHLKVSNIVVI 213
K+S + +I+Y VLH+KV N+VV+
Sbjct: 368 TKYSAAITSIKYTVLHVKVENMVVL 442
>CD392487
Length = 384
Score = 153 bits (386), Expect = 1e-37
Identities = 72/81 (88%), Positives = 77/81 (94%)
Frame = -3
Query: 244 IGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYD 303
IGLPAKAKVK +HGDAPF ELC+HCEKEAVNVSLGNLL+YPFVR+GLVNKTL+LKGGYYD
Sbjct: 310 IGLPAKAKVKTQHGDAPFAELCSHCEKEAVNVSLGNLLTYPFVRDGLVNKTLSLKGGYYD 131
Query: 304 FVKGSFELWSLQFDLASSFSV 324
FVKGSFELW LQF LASSFSV
Sbjct: 130 FVKGSFELWGLQFGLASSFSV 68
>BE331068 weakly similar to GP|8954289|gb| carbonic anhydrase {Vigna
radiata}, partial (32%)
Length = 517
Score = 139 bits (351), Expect = 1e-33
Identities = 80/171 (46%), Positives = 113/171 (65%), Gaps = 6/171 (3%)
Frame = +2
Query: 3 TSTINGFYLSSISPAKTSLKRVTLRPLV---VASASSPSSSSSSSFPSLIQDRPVFAAPA 59
+S+I + LS +SPAKTSL + TL + + + S PS SS SFP++IQ+ P+ P+
Sbjct: 5 SSSIGEWRLSCMSPAKTSLSKATLGACICRTIDTPSCPSGWSSGSFPAVIQNEPILTTPS 184
Query: 60 PII-PTL--DMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEA 116
++ PT+ +M + E+ I+ LQKLLREK + T+ +KVE IT L T+SS+ IPSS++
Sbjct: 185 DMVTPTVRRNMARK*EQTIK*LQKLLREKIGRQTTSTKKVEHITTSL*TSSSNNIPSSQS 364
Query: 117 SDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLD 167
S RIK F++F K KY NPALYGEL K +++F SDS+V PSHV D
Sbjct: 365 SYRIKANFIHFNKYKYYNNPALYGELTKYHRRKFIIFTYSDSQVWPSHVRD 517
>TC203698 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (21%)
Length = 1348
Score = 129 bits (323), Expect = 2e-30
Identities = 60/68 (88%), Positives = 64/68 (93%)
Frame = +3
Query: 257 GDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQF 316
GDAPF ELC+HCEKE VNVSLGNLL+YPFVR+GLVNKTL+LKGGYYDFVKGSFELW LQF
Sbjct: 969 GDAPFAELCSHCEKEFVNVSLGNLLTYPFVRDGLVNKTLSLKGGYYDFVKGSFELWGLQF 1148
Query: 317 DLASSFSV 324
LASSFSV
Sbjct: 1149 GLASSFSV 1172
>BE609533 similar to GP|8954289|gb|A carbonic anhydrase {Vigna radiata},
partial (24%)
Length = 244
Score = 113 bits (282), Expect = 1e-25
Identities = 53/81 (65%), Positives = 63/81 (77%)
Frame = +2
Query: 238 IEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLAL 297
IE+WV IGLPA+A VK HGDAP +LC+HCEKEA NVSLGNLL+YPF +G+ N TL+L
Sbjct: 2 IEEWVKIGLPAQAMVKTPHGDAPSADLCSHCEKEADNVSLGNLLTYPFDTDGVENTTLSL 181
Query: 298 KGGYYDFVKGSFELWSLQFDL 318
GYYD+ KGS +L LQF L
Sbjct: 182 NRGYYDYAKGSSDL*GLQFGL 244
>TC223179 weakly similar to UP|O81875 (O81875) Carbonate dehydratase-like
protein, partial (62%)
Length = 434
Score = 108 bits (271), Expect = 3e-24
Identities = 52/108 (48%), Positives = 68/108 (62%)
Frame = +2
Query: 209 NIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHC 268
NI+VIGHS CGGI+ L+ D FI+ WV G A+ K KA + F E C HC
Sbjct: 5 NILVIGHSCCGGIRALMGMQ-DDDVEKSFIKSWVIAGKNARKKAKAAASNLSFDEQCKHC 181
Query: 269 EKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQF 316
EKE++N SL NLL+YP++ E + N L++ GGYYDF SFE W+L +
Sbjct: 182 EKESINHSLLNLLTYPWIEEKVANGELSIHGGYYDFTDCSFEKWTLDY 325
>AW132257 similar to GP|8569250|pdb| Chain A The X-Ray Crystallographic
Structure Of Beta Carbonic Anhydrase From The C3 Dicot
Pisum, partial (24%)
Length = 337
Score = 108 bits (270), Expect = 3e-24
Identities = 57/111 (51%), Positives = 75/111 (67%), Gaps = 3/111 (2%)
Frame = +2
Query: 72 EEAIEE---LQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFK 128
+E I+E L +LL EK EL AA +++++TA+L + S + F++FK
Sbjct: 14 DEIIQEHTSLTRLLSEKAELGGRAAAQIKELTAELDADVRSRVTRRRGS---RNRFIHFK 184
Query: 129 KEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNV 179
E ++KNP LYGELA GQSP MVFACSDSRVCPSH+LD QPGEAF+VR +
Sbjct: 185 NEXFEKNPYLYGELAIGQSPIVMVFACSDSRVCPSHILDLQPGEAFMVRTL 337
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,202,869
Number of Sequences: 63676
Number of extensions: 211311
Number of successful extensions: 2520
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 1970
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2269
length of query: 324
length of database: 12,639,632
effective HSP length: 97
effective length of query: 227
effective length of database: 6,463,060
effective search space: 1467114620
effective search space used: 1467114620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0201.15


