

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
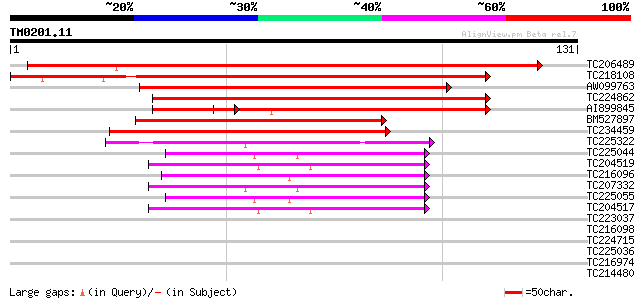
Query= TM0201.11
(131 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206489 similar to UP|Q9XGG7 (Q9XGG7) Nodulin26-like major intr... 178 6e-46
TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete 135 4e-33
AW099763 similar to PIR|JQ2285|JQ22 nodulin-26 - soybean, partia... 122 5e-29
TC224862 similar to UP|Q6KAW0 (Q6KAW0) Nod26-like major intrinsi... 71 1e-13
AI899845 weakly similar to GP|13447789|gb| NOD26-like membrane i... 53 3e-10
BM527897 56 6e-09
TC234459 similar to UP|Q6QIL3 (Q6QIL3) NIP3, partial (41%) 55 9e-09
TC225322 similar to UP|TI41_ARATH (O82316) Probable aquaporin TI... 53 4e-08
TC225044 homologue to UP|O22339 (O22339) Aquaporin-like transmem... 42 1e-04
TC204519 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic... 41 2e-04
TC216096 homologue to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (P... 41 2e-04
TC207332 similar to UP|TIPA_PHAVU (P23958) Probable aquaporin TI... 40 3e-04
TC225055 homologue to UP|Q9LLL9 (Q9LLL9) Plasma membrane aquapor... 39 5e-04
TC204517 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic... 39 7e-04
TC223037 similar to UP|Q8H1Z5 (Q8H1Z5) Tonoplast intrinsic prote... 39 0.001
TC216098 similar to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (Pla... 38 0.001
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 37 0.002
TC225036 similar to UP|O65357 (O65357) Aquaporin 2, partial (92%) 37 0.002
TC216974 homologue to UP|Q39822 (Q39822) Pip1 protein, complete 37 0.002
TC214480 homologue to UP|Q39822 (Q39822) Pip1 protein, complete 37 0.002
>TC206489 similar to UP|Q9XGG7 (Q9XGG7) Nodulin26-like major intrinsic
protein, partial (86%)
Length = 1169
Score = 178 bits (452), Expect = 6e-46
Identities = 88/120 (73%), Positives = 103/120 (85%), Gaps = 1/120 (0%)
Frame = +1
Query: 5 SGSNEVILNVNNETSKKCD-SIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 63
+GS++V+LNVN + KKCD S +DCVPLLQKLVAEV+GTYFLIFAGCASVVVNL+ DKV
Sbjct: 124 NGSHQVVLNVNGDAPKKCDDSANQDCVPLLQKLVAEVVGTYFLIFAGCASVVVNLDKDKV 303
Query: 64 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
VT PGI+IVWGL VMVLVYS+GHISGAHFNPAVT+AHATTK FP+KQV + + VG +
Sbjct: 304 VTQPGISIVWGLTVMVLVYSVGHISGAHFNPAVTIAHATTKRFPLKQVPAYVIAQVVGAT 483
>TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete
Length = 1122
Score = 135 bits (341), Expect = 4e-33
Identities = 72/117 (61%), Positives = 88/117 (74%), Gaps = 6/117 (5%)
Frame = +3
Query: 1 MADYSG---SNEVILNVNNETSK---KCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASV 54
MADYS S EV++NV TS+ + DS+ VP LQKLVAE +GTYFLIFAGCAS+
Sbjct: 93 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVS--VPFLQKLVAEAVGTYFLIFAGCASL 266
Query: 55 VVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
VVN N ++T PGIAIVWGL + VLVY++GHISG HFNPAVT+A A+T+ FP+ QV
Sbjct: 267 VVNENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQV 437
>AW099763 similar to PIR|JQ2285|JQ22 nodulin-26 - soybean, partial (34%)
Length = 472
Score = 122 bits (306), Expect = 5e-29
Identities = 59/72 (81%), Positives = 65/72 (89%)
Frame = +2
Query: 31 PLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGA 90
PLL KLVAEV+GTYFLIFAGCASVV NL+ DKVVT PGI+IVWGL VMVLVYS GHISGA
Sbjct: 2 PLLHKLVAEVVGTYFLIFAGCASVVENLDKDKVVTQPGISIVWGLTVMVLVYSFGHISGA 181
Query: 91 HFNPAVTVAHAT 102
HF PAVT+AH++
Sbjct: 182 HFYPAVTIAHSS 217
>TC224862 similar to UP|Q6KAW0 (Q6KAW0) Nod26-like major intrinsic protein,
partial (79%)
Length = 1192
Score = 71.2 bits (173), Expect = 1e-13
Identities = 33/78 (42%), Positives = 52/78 (66%)
Frame = +1
Query: 34 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 93
+K+ AEV+GT+ L+F G S ++ ++ +V+ G ++ GL V V++YSIGHISGAH N
Sbjct: 274 RKVFAEVIGTFLLVFVGSGSAGLSKIDESMVSKLGASLAGGLIVTVMIYSIGHISGAHMN 453
Query: 94 PAVTVAHATTKSFPVKQV 111
PAV++A + P Q+
Sbjct: 454 PAVSLAFTAVRHLPWPQL 507
Score = 36.6 bits (83), Expect = 0.003
Identities = 23/74 (31%), Positives = 39/74 (52%)
Frame = +1
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
+Q L+ E++ TY ++F S+ V +++ L G+A+ G +V + G ISG
Sbjct: 619 IQALIMEMVSTYTMVFI---SMAVATDSNATGQLSGVAV--GSSVCIASIVAGPISGGSM 783
Query: 93 NPAVTVAHATTKSF 106
NPA T+ A S+
Sbjct: 784 NPARTLGPAIATSY 825
>AI899845 weakly similar to GP|13447789|gb| NOD26-like membrane integral
protein ZmNIP2-2 {Zea mays}, partial (31%)
Length = 430
Score = 52.8 bits (125), Expect(2) = 3e-10
Identities = 28/68 (41%), Positives = 41/68 (60%), Gaps = 4/68 (5%)
Frame = +1
Query: 48 FAGCASVVVNLN----NDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATT 103
F C VV L ++++V+ G ++ GL V V++YSIGHISGAH NPAV++A
Sbjct: 169 FCWCLWEVVLLGLSKIDERMVSKLGASLAGGLIVTVMIYSIGHISGAHMNPAVSLAFTAV 348
Query: 104 KSFPVKQV 111
+ P Q+
Sbjct: 349 RHLPWPQL 372
Score = 26.9 bits (58), Expect(2) = 3e-10
Identities = 9/20 (45%), Positives = 16/20 (80%)
Frame = +3
Query: 34 QKLVAEVLGTYFLIFAGCAS 53
+K++AE++GT+ L+F G S
Sbjct: 138 RKVLAEIIGTFLLVFVGSGS 197
>BM527897
Length = 433
Score = 55.8 bits (133), Expect = 6e-09
Identities = 27/58 (46%), Positives = 39/58 (66%)
Frame = +2
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHI 87
VPL +K+ AE +GT+ L+FAG A+ +VN + TL G A GLAVM+++ + GHI
Sbjct: 260 VPLARKIGAEFIGTFILMFAGTAAAIVNQKTNGSETLIGCAATTGLAVMIVILATGHI 433
>TC234459 similar to UP|Q6QIL3 (Q6QIL3) NIP3, partial (41%)
Length = 460
Score = 55.1 bits (131), Expect = 9e-09
Identities = 27/65 (41%), Positives = 40/65 (61%)
Frame = +3
Query: 24 SIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYS 83
S+ +PL +K+ AE +GT+ L+FA + +VN TL G A GLAVM++++S
Sbjct: 264 SLPSPHIPLAKKIGAEFIGTFILMFAAIGTAIVNQKTHGSETLIGCAAANGLAVMIIIFS 443
Query: 84 IGHIS 88
GHIS
Sbjct: 444 TGHIS 458
>TC225322 similar to UP|TI41_ARATH (O82316) Probable aquaporin TIP4.1
(Tonoplast intrinsic protein 4.1) (Epsilon-tonoplast
intrinsic protein) (Epsilon-TIP), partial (98%)
Length = 1378
Score = 53.1 bits (126), Expect = 4e-08
Identities = 30/78 (38%), Positives = 44/78 (55%), Gaps = 2/78 (2%)
Frame = +2
Query: 23 DSIEEDCVPLLQKLVAEVLGTYFLIFAGCAS--VVVNLNNDKVVTLPGIAIVWGLAVMVL 80
++ + DC+ Q L+ E + T+ +F G S VV L D +V L +A+ L V V+
Sbjct: 575 EATQPDCI---QALIVEFIATFLFVFVGVGSSMVVDKLGGDALVGLFAVAVAHALVVAVM 745
Query: 81 VYSIGHISGAHFNPAVTV 98
+ S HISG H NPAVT+
Sbjct: 746 I-SAAHISGGHLNPAVTL 796
>TC225044 homologue to UP|O22339 (O22339) Aquaporin-like transmembrane
channel protein, complete
Length = 1235
Score = 41.6 bits (96), Expect = 1e-04
Identities = 24/64 (37%), Positives = 36/64 (55%), Gaps = 3/64 (4%)
Frame = +2
Query: 37 VAEVLGTYFLIFAGCASVV-VNLNNDKVVT--LPGIAIVWGLAVMVLVYSIGHISGAHFN 93
+AE + T+ ++ +V+ VN ++ K T + GIA +G + LVY ISG H N
Sbjct: 203 IAEFVATFLFLYITILTVMGVNRSSSKCATVGIQGIAWAFGGMIFALVYCTAGISGGHIN 382
Query: 94 PAVT 97
PAVT
Sbjct: 383 PAVT 394
>TC204519 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
partial (97%)
Length = 989
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/70 (35%), Positives = 36/70 (50%), Gaps = 5/70 (7%)
Frame = +1
Query: 33 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 87
++ +AE + T +FAG S + L +D + G +AI G A+ V V +I
Sbjct: 112 IKAYIAEFISTLLFVFAGVGSAIAYAKLTSDAALDPTGLVAVAICHGFALFVAVSVGANI 291
Query: 88 SGAHFNPAVT 97
SG H NPAVT
Sbjct: 292 SGGHVNPAVT 321
>TC216096 homologue to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (Plasma
membrane intrinsic protein 3) (Salt-stress induced major
intrinsis protein), partial (97%)
Length = 1043
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/65 (38%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Frame = +1
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
L+AE + T ++ A+V+ + V L GIA +G + VLVY ISG H
Sbjct: 100 LIAEFIATLLFLYVTVATVIGHKKQTGPCDGVGLLGIAWAFGGMIFVLVYCTAGISGGHI 279
Query: 93 NPAVT 97
NPAVT
Sbjct: 280 NPAVT 294
>TC207332 similar to UP|TIPA_PHAVU (P23958) Probable aquaporin TIP-type alpha
(Tonoplast intrinsic protein alpha) (Alpha TIP), partial
(54%)
Length = 540
Score = 40.0 bits (92), Expect = 3e-04
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 5/70 (7%)
Frame = +1
Query: 33 LQKLVAEVLGTYFLIFAGCAS--VVVNLNNDKVVT---LPGIAIVWGLAVMVLVYSIGHI 87
++ +AE + T+ +FAG S +V + D + L +A+ G A+ V + H+
Sbjct: 181 MRATLAEFVSTFIFVFAGEGSGLALVKIYQDSAFSAGELLAVALAHGFALFAAVSASMHV 360
Query: 88 SGAHFNPAVT 97
SG H NPAVT
Sbjct: 361 SGGHVNPAVT 390
>TC225055 homologue to UP|Q9LLL9 (Q9LLL9) Plasma membrane aquaporin, complete
Length = 1148
Score = 39.3 bits (90), Expect = 5e-04
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Frame = +3
Query: 37 VAEVLGTYFLIFAGCASVV-VNLNNDKV--VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 93
+AE + T+ ++ +V+ VN + K V + GIA +G + LVY ISG H N
Sbjct: 207 IAEFVATFLFLYITILTVMGVNRSPSKCASVGIQGIAWAFGGMIFALVYCTAGISGGHIN 386
Query: 94 PAVT 97
PAVT
Sbjct: 387 PAVT 398
>TC204517 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
complete
Length = 1016
Score = 38.9 bits (89), Expect = 7e-04
Identities = 25/70 (35%), Positives = 35/70 (49%), Gaps = 5/70 (7%)
Frame = +3
Query: 33 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 87
++ +AE T +FAG S + L +D + G +AI G A+ V V +I
Sbjct: 120 IKAYIAEFHSTLLFVFAGVGSAIAYGKLTSDAALDPAGLLAVAICHGFALFVAVSVGANI 299
Query: 88 SGAHFNPAVT 97
SG H NPAVT
Sbjct: 300 SGGHVNPAVT 329
>TC223037 similar to UP|Q8H1Z5 (Q8H1Z5) Tonoplast intrinsic protein
bobTIP26-2, partial (76%)
Length = 723
Score = 38.5 bits (88), Expect = 0.001
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Frame = +3
Query: 37 VAEVLGTYFLIFAGC-ASVVVN-LNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNP 94
++E + T +FAG +SV VN L DK L A+ A+ V V +ISG H NP
Sbjct: 138 LSEFISTLIFVFAGSGSSVAVNKLTVDKPSALVVAAVAHAFALFVAVSVSTNISGGHVNP 317
Query: 95 AVT 97
AVT
Sbjct: 318 AVT 326
>TC216098 similar to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (Plasma membrane
intrinsic protein 3) (Salt-stress induced major
intrinsis protein), partial (81%)
Length = 838
Score = 38.1 bits (87), Expect = 0.001
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Frame = +2
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
L+AE + + ++ A+++ + V L GIA +G + VLVY ISG H
Sbjct: 260 LIAEFIASLLFLYVTVATIIGHKKQTGPCDGVGLLGIAWSFGGMIFVLVYCTAGISGGHI 439
Query: 93 NPAVT 97
NPAVT
Sbjct: 440 NPAVT 454
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 37.4 bits (85), Expect = 0.002
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Frame = +1
Query: 33 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGI---AIVWGLAVMVLVYSIGHI 87
L+ +AE + T+ +FAG S + L ++ T G+ +I A+ V V +I
Sbjct: 559 LKAALAEFISTFIFVFAGSGSGIAYNKLTDNGAATPAGLISASIAHAFALFVAVSVGANI 738
Query: 88 SGAHFNPAVT 97
SG H NPAVT
Sbjct: 739 SGGHVNPAVT 768
Score = 26.6 bits (57), Expect = 3.6
Identities = 20/66 (30%), Positives = 30/66 (45%)
Frame = +1
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPA 95
LV E++ T+ L++ A+ + + + P I G V + G SGA NPA
Sbjct: 931 LVLEIVMTFGLVYTVYATAIDPKKGNLGIIAP---IAIGFIVGANILLGGAFSGAAMNPA 1101
Query: 96 VTVAHA 101
VT A
Sbjct: 1102VTFGPA 1119
>TC225036 similar to UP|O65357 (O65357) Aquaporin 2, partial (92%)
Length = 1610
Score = 37.4 bits (85), Expect = 0.002
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 7/69 (10%)
Frame = +3
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKVVTLP-------GIAIVWGLAVMVLVYSIGHIS 88
L+AE + T ++ +V+ + P GIA +G + VLVY IS
Sbjct: 597 LIAEFVATLLFLYVTILTVIGYNHQTATAAEPCSGVGVLGIAWAFGGMIFVLVYCTAGIS 776
Query: 89 GAHFNPAVT 97
G H NPAVT
Sbjct: 777 GGHINPAVT 803
>TC216974 homologue to UP|Q39822 (Q39822) Pip1 protein, complete
Length = 1254
Score = 37.4 bits (85), Expect = 0.002
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 9/71 (12%)
Frame = +3
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---------VTLPGIAIVWGLAVMVLVYSIGH 86
L+AE + T ++ +V+ + V V + GIA +G + +LVY
Sbjct: 210 LIAEFIATMLFLYITVLTVIGYKSQSDVKAGGDVCGGVGILGIAWAFGGMIFILVYCTAG 389
Query: 87 ISGAHFNPAVT 97
ISG H NPAVT
Sbjct: 390 ISGGHINPAVT 422
>TC214480 homologue to UP|Q39822 (Q39822) Pip1 protein, complete
Length = 1240
Score = 37.4 bits (85), Expect = 0.002
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 9/71 (12%)
Frame = +1
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---------VTLPGIAIVWGLAVMVLVYSIGH 86
L+AE + T ++ +V+ + V V + GIA +G + +LVY
Sbjct: 214 LIAEFIATMLFLYITVLTVIGYKSQSDVKAGGDVCGGVGILGIAWAFGGMIFILVYCTAG 393
Query: 87 ISGAHFNPAVT 97
ISG H NPAVT
Sbjct: 394 ISGGHINPAVT 426
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,033,259
Number of Sequences: 63676
Number of extensions: 73043
Number of successful extensions: 408
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 406
length of query: 131
length of database: 12,639,632
effective HSP length: 86
effective length of query: 45
effective length of database: 7,163,496
effective search space: 322357320
effective search space used: 322357320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0201.11


