

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
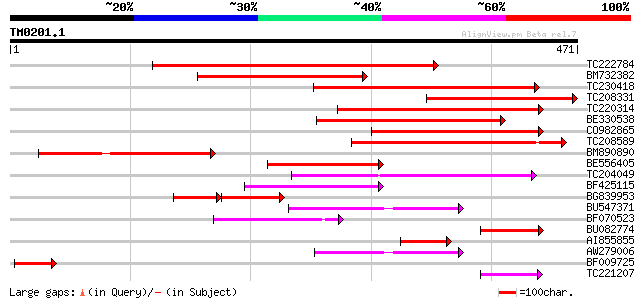
Query= TM0201.1
(471 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222784 similar to UP|ALA8_ARATH (Q9LK90) Potential phospholipi... 461 e-130
BM732382 similar to SP|Q9LK90|ALA Potential phospholipid-transpo... 258 3e-69
TC230418 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential phos... 257 9e-69
TC208331 weakly similar to UP|ALA8_ARATH (Q9LK90) Potential phos... 226 2e-59
TC220314 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential phos... 216 2e-56
BE330538 similar to SP|Q9SGG3|ALA Potential phospholipid-transpo... 197 1e-50
CO982865 187 7e-48
TC208589 similar to GB|AAF79467.1|8778459|AC022492 F1L3.21 {Arab... 180 1e-45
BM890890 similar to SP|Q9LI83|ALA Potential phospholipid-transpo... 152 2e-37
BE556405 similar to SP|Q9SX33|ALA9 Potential phospholipid-transp... 150 2e-36
TC204049 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to ch... 126 2e-29
BF425115 similar to SP|P98204|ALA Phospholipid-transporting ATPa... 95 6e-20
BG839953 65 1e-18
BU547371 weakly similar to SP|P98204|ALA Phospholipid-transporti... 72 4e-13
BF070523 similar to GP|16930515|gb AT5g44240/MLN1_17 {Arabidopsi... 66 3e-11
BU082774 weakly similar to SP|Q9SX33|ALA9 Potential phospholipid... 62 6e-10
AI855855 similar to SP|Q9SAF5|ALAB Potential phospholipid-transp... 61 1e-09
AW279006 weakly similar to SP|P98204|ALA1 Phospholipid-transport... 60 2e-09
BF009725 similar to SP|P98204|ALA Phospholipid-transporting ATPa... 51 1e-06
TC221207 50 2e-06
>TC222784 similar to UP|ALA8_ARATH (Q9LK90) Potential
phospholipid-transporting ATPase 8 (Aminophospholipid
flippase 8) , partial (20%)
Length = 716
Score = 461 bits (1186), Expect = e-130
Identities = 220/238 (92%), Positives = 232/238 (97%)
Frame = +3
Query: 119 VSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQ 178
++CASVICCRSSPKQKARVT+LVKLGTGKT LSIGDGANDVGMLQEA IGVGISGAEGMQ
Sbjct: 3 INCASVICCRSSPKQKARVTKLVKLGTGKTTLSIGDGANDVGMLQEADIGVGISGAEGMQ 182
Query: 179 AVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQ 238
AVMASDFAIAQFRFLERLLLVHGHWCYRRIS+MICYFFYKNIAFGFTLFWFEAYASFSGQ
Sbjct: 183 AVMASDFAIAQFRFLERLLLVHGHWCYRRISMMICYFFYKNIAFGFTLFWFEAYASFSGQ 362
Query: 239 PAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWM 298
AYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLK+P+LYLEGVED LFSWPRI+GWM
Sbjct: 363 AAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKHPYLYLEGVEDILFSWPRILGWM 542
Query: 299 LNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYF 356
LNGV+SSL IFFLTTNS++NQAFRRDG+VVDFEILGVTMY+ VVWTVNCQMALSINYF
Sbjct: 543 LNGVLSSLVIFFLTTNSVLNQAFRRDGKVVDFEILGVTMYTCVVWTVNCQMALSINYF 716
>BM732382 similar to SP|Q9LK90|ALA Potential phospholipid-transporting ATPase
8 (EC 3.6.3.1) (Aminophospholipid flippase 8)., partial
(11%)
Length = 429
Score = 258 bits (660), Expect = 3e-69
Identities = 120/141 (85%), Positives = 130/141 (92%)
Frame = +2
Query: 157 NDVGMLQEAHIGVGISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFF 216
NDVGMLQEA IG+GISG EGMQAVM+SD AIAQFRFLERLLLVHGHWCYRRIS MICYFF
Sbjct: 2 NDVGMLQEADIGIGISGVEGMQAVMSSDIAIAQFRFLERLLLVHGHWCYRRISSMICYFF 181
Query: 217 YKNIAFGFTLFWFEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKY 276
YKNIAFGFTLF++E YASFSGQ AYNDWY+S YNVFFTSLPVIALGVFDQDVSA+LCLK+
Sbjct: 182 YKNIAFGFTLFFYEIYASFSGQAAYNDWYLSLYNVFFTSLPVIALGVFDQDVSARLCLKF 361
Query: 277 PFLYLEGVEDTLFSWPRIIGW 297
P LY EGV++ LFSW RI+GW
Sbjct: 362 PLLYQEGVQNVLFSWKRILGW 424
>TC230418 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential
phospholipid-transporting ATPase 9 (Aminophospholipid
flippase 9) , partial (19%)
Length = 1062
Score = 257 bits (656), Expect = 9e-69
Identities = 116/188 (61%), Positives = 145/188 (76%)
Frame = +1
Query: 253 FTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLT 312
FTSLPVIALGVFDQDVS+KLCLK+P LY EG ++ LFSW RIIGW LNGV++S +FF
Sbjct: 1 FTSLPVIALGVFDQDVSSKLCLKFPLLYQEGTQNILFSWKRIIGWALNGVVTSAIVFFFC 180
Query: 313 TNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWY 372
S+ QAFR+ G+V+ E+LG TMY+ VVW VNCQMALSI+YFT+IQH FIWGSI FW+
Sbjct: 181 IRSMEYQAFRKGGEVMGLEVLGATMYTCVVWVVNCQMALSISYFTYIQHIFIWGSILFWH 360
Query: 373 VFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMY 432
+FLL YG + P+ S+TAY F+EA AP+ +W+ T L+++ LLPYF Y + Q RF PMY
Sbjct: 361 IFLLAYGAIDPSFSTTAYKGFIEALAPAPFFWIITFLILIASLLPYFIYASIQMRFFPMY 540
Query: 433 HDIIQRKR 440
H +IQ R
Sbjct: 541 HQMIQWMR 564
>TC208331 weakly similar to UP|ALA8_ARATH (Q9LK90) Potential
phospholipid-transporting ATPase 8 (Aminophospholipid
flippase 8) , partial (8%)
Length = 935
Score = 226 bits (575), Expect = 2e-59
Identities = 107/126 (84%), Positives = 117/126 (91%), Gaps = 1/126 (0%)
Frame = +3
Query: 347 CQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLA 406
CQMALSINYFTWIQHFFIWGSI FWYVF+LVYGYLSPAIS+TAY VFVEACAPS +YWL
Sbjct: 3 CQMALSINYFTWIQHFFIWGSIAFWYVFVLVYGYLSPAISTTAYRVFVEACAPSGLYWLV 182
Query: 407 TVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGFEVEIS-DELPTQVQGKLMHMRE 465
T+LVVVCVLLPYF+YR+FQSRFLPMYHDIIQRK+VEG EV +S DELP QVQ KL+H+RE
Sbjct: 183 TLLVVVCVLLPYFSYRSFQSRFLPMYHDIIQRKQVEGHEVGLSDDELPKQVQDKLLHLRE 362
Query: 466 RLKQRE 471
RLKQRE
Sbjct: 363 RLKQRE 380
>TC220314 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential
phospholipid-transporting ATPase 9 (Aminophospholipid
flippase 9) , partial (15%)
Length = 647
Score = 216 bits (549), Expect = 2e-56
Identities = 99/171 (57%), Positives = 124/171 (71%)
Frame = +2
Query: 273 CLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEI 332
CLK+P LY EGV++ LFSW RI+ WMLNG IS+L IFF T ++ QAF +G+ +I
Sbjct: 5 CLKFPLLYQEGVQNVLFSWRRILSWMLNGFISALIIFFFCTKAMELQAFDVEGRTAGKDI 184
Query: 333 LGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMV 392
LG MY+ VVW VN QMAL+++YFT IQHFFIWGSI WY+FL+VYG + P S+ AY V
Sbjct: 185 LGAAMYTCVVWVVNLQMALAVSYFTMIQHFFIWGSILLWYLFLVVYGAMPPHFSTNAYKV 364
Query: 393 FVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEG 443
FVEA APS YW+ T VV+ L+PYF+Y A + RF PMYH+I+Q R EG
Sbjct: 365 FVEALAPSPSYWIVTFFVVISTLIPYFSYAAIRMRFFPMYHEIVQWIRYEG 517
>BE330538 similar to SP|Q9SGG3|ALA Potential phospholipid-transporting ATPase
5 (EC 3.6.3.1) (Aminophospholipid flippase 5)., partial
(12%)
Length = 472
Score = 197 bits (500), Expect = 1e-50
Identities = 84/157 (53%), Positives = 118/157 (74%)
Frame = +1
Query: 256 LPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTTNS 315
LPVI+LGVF+QDV +++CL++P LY +G ++ F W RI+GWM NG+ +SL IFFL
Sbjct: 1 LPVISLGVFEQDVPSEVCLQFPALYQQGPKNLFFDWYRILGWMGNGLYASLIIFFLIVTI 180
Query: 316 IINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFL 375
+QAFR DGQV D +G TM++ ++WTVNCQ+AL++++FTWIQH F+WGSI WY+FL
Sbjct: 181 FYDQAFRADGQVADMAAVGTTMFTCIIWTVNCQIALTMSHFTWIQHLFVWGSIATWYIFL 360
Query: 376 LVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVV 412
+YG LSP S +AY + VE+ P+ +YW+ T+LV V
Sbjct: 361 SLYGMLSPEYSKSAYQILVESLGPAPIYWVTTLLVTV 471
>CO982865
Length = 730
Score = 187 bits (476), Expect = 7e-48
Identities = 85/143 (59%), Positives = 107/143 (74%)
Frame = -1
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV+S+ IFF N + NQAFR+ G+V D E+LG TMY+ VVW VN QMALSI+YFT+IQ
Sbjct: 730 GVLSATIIFFFCINGMENQAFRKAGEVADLEVLGATMYTCVVWVVNSQMALSISYFTYIQ 551
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H FIWG I FWY+FLLVYG + P++S+TAY V +EACAP+ YWL T+LV+V LLPYF
Sbjct: 550 HLFIWGGILFWYIFLLVYGTMDPSLSTTAYKVLIEACAPAPSYWLITLLVLVASLLPYFA 371
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
Y + Q RF P +H +IQ R +G
Sbjct: 370 YASIQMRFFPTFHQMIQWIRNDG 302
>TC208589 similar to GB|AAF79467.1|8778459|AC022492 F1L3.21 {Arabidopsis
thaliana;} , partial (14%)
Length = 843
Score = 180 bits (457), Expect = 1e-45
Identities = 84/179 (46%), Positives = 120/179 (66%), Gaps = 1/179 (0%)
Frame = +1
Query: 285 EDTLFSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWT 344
++ F W RI+GWM NG+ SSL IFFL +QAFR +GQ D +G TM++ ++W
Sbjct: 1 KNLFFDWYRILGWMGNGLYSSLIIFFLVIIIFYDQAFRANGQTTDMAAVGTTMFTCIIWA 180
Query: 345 VNCQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYW 404
VNCQ+AL++++FTWIQH F+WGSI WYVFLL+YG L P S +AY + VE AP+ +YW
Sbjct: 181 VNCQIALTMSHFTWIQHLFVWGSITTWYVFLLLYGMLPPQYSKSAYQLLVEVLAPAPIYW 360
Query: 405 LATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGFEVEISDE-LPTQVQGKLMH 462
AT+LV + +LPY + +FQ F PM H IIQ ++ ++ +I D+ + T+ + K H
Sbjct: 361 AATLLVTIACVLPYLAHISFQRCFNPMDHHIIQ--EIKYYKKDIEDQHMWTRERSKARH 531
>BM890890 similar to SP|Q9LI83|ALA Potential phospholipid-transporting ATPase
10 (EC 3.6.3.1) (Aminophospholipid flippase 10).,
partial (10%)
Length = 425
Score = 152 bits (385), Expect = 2e-37
Identities = 82/147 (55%), Positives = 103/147 (69%)
Frame = +1
Query: 25 YACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKASLESIKKQISEGISQVKSAKESS 84
+ACSLLRQ MK+I+I+ D+P+ SLEK DK A A S+ Q++ G + +
Sbjct: 1 FACSLLRQGMKQIIISSDTPETKSLEKVEDKSAAAAAVKVSVIHQLTNGKELL------A 162
Query: 85 NTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSCASVICCRSSPKQKARVTRLVKLG 144
+D+ + A LIIDGKSL Y+L +++ F LA CASVICCRSSPKQKA VTRLVK+
Sbjct: 163 ESDENSEALALIIDGKSLTYALEDDVKDLFLTLAAGCASVICCRSSPKQKALVTRLVKVK 342
Query: 145 TGKTILSIGDGANDVGMLQEAHIGVGI 171
TG T L+IGDGANDVGMLQEA IG+GI
Sbjct: 343 TGSTTLAIGDGANDVGMLQEADIGIGI 423
>BE556405 similar to SP|Q9SX33|ALA9 Potential phospholipid-transporting
ATPase 9 (EC 3.6.3.1) (Aminophospholipid flippase 9).,
partial (8%)
Length = 289
Score = 150 bits (378), Expect = 2e-36
Identities = 68/96 (70%), Positives = 81/96 (83%)
Frame = +2
Query: 215 FFYKNIAFGFTLFWFEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCL 274
FFYKNI FGFTLF +E YASFSGQPAYNDW++S YNVFF+SLP IALGVFDQDVSA+ CL
Sbjct: 2 FFYKNITFGFTLFLYEEYASFSGQPAYNDWFLSLYNVFFSSLPEIALGVFDQDVSARNCL 181
Query: 275 KYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFF 310
++P LY EGV++ LFS +I WMLNG IS++ I+F
Sbjct: 182 RFPMLYQEGVQNVLFSCRQIFSWMLNGFISAIIIYF 289
>TC204049 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to chromaffin
granule ATPase II homolog, partial (24%)
Length = 1234
Score = 126 bits (316), Expect = 2e-29
Identities = 73/204 (35%), Positives = 105/204 (50%), Gaps = 1/204 (0%)
Frame = +2
Query: 235 FSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRI 294
FSGQ Y+DW+ S YNV FT+LPVI +G+FD+DVS+ L KYP LY+EG+ + F W +
Sbjct: 14 FSGQRFYDDWFQSLYNVIFTALPVIIVGLFDKDVSSSLSKKYPQLYMEGIRNVFFKWKVV 193
Query: 295 IGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSIN 354
W V SL IFF +S A G++ + ++ VV TVN ++ + N
Sbjct: 194 AIWAFFSVYQSL-IFFYFVSSTNLSAKNSAGKIFGLWDVSTMAFTCVVITVNLRLLMICN 370
Query: 355 YFTWIQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVY-WLATVLVVVC 413
T + + GSI W++F+ +Y +S + FV S Y ++ LV V
Sbjct: 371 SITRWHYISVGGSILAWFLFIFIYSGISTPYDRQENIYFVIYVLMSTFYFYVMLFLVPVA 550
Query: 414 VLLPYFTYRAFQSRFLPMYHDIIQ 437
L F Y+ Q F P + IIQ
Sbjct: 551 ALFCDFVYQGVQRWFFPYDYQIIQ 622
>BF425115 similar to SP|P98204|ALA Phospholipid-transporting ATPase 1 (EC
3.6.3.1) (Aminophospholipid flippase 1). [Mouse-ear
cress], partial (10%)
Length = 384
Score = 95.1 bits (235), Expect = 6e-20
Identities = 42/115 (36%), Positives = 68/115 (58%)
Frame = +2
Query: 196 LLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAYNDWYMSFYNVFFTS 255
LLL+HGHW Y+R+ MI Y FY+N F LFW+ + +F+ A N+W Y++ +++
Sbjct: 8 LLLIHGHWNYQRLGYMIIYNFYRNAIFVLVLFWYVLFTAFTLTTAINEWSSVLYSIIYSA 187
Query: 256 LPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFF 310
P I +G+ D+D+S + LKYP LY G+ ++ M + + S+A+FF
Sbjct: 188 FPTIGVGILDKDLSKRTLLKYPQLYGAGLRQEAYNKKLFWLAMADTLWQSIAVFF 352
>BG839953
Length = 730
Score = 65.5 bits (158), Expect(2) = 1e-18
Identities = 29/52 (55%), Positives = 35/52 (66%)
Frame = +1
Query: 177 MQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFW 228
+QAVMASDFA+ FRF LLL+H HW Y+R+ MI Y FY+N LFW
Sbjct: 124 VQAVMASDFAMGHFRFSVPLLLIHSHWNYQRLGYMILYNFYRNAVLVLVLFW 279
Score = 45.4 bits (106), Expect(2) = 1e-18
Identities = 21/40 (52%), Positives = 26/40 (64%)
Frame = +3
Query: 137 VTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEG 176
+ LVK T L+IGDGANDV M+Q +G+G SG EG
Sbjct: 6 IVALVKNRTSDMTLAIGDGANDVSMIQMVDVGIGFSGQEG 125
>BU547371 weakly similar to SP|P98204|ALA Phospholipid-transporting ATPase 1
(EC 3.6.3.1) (Aminophospholipid flippase 1). [Mouse-ear
cress], partial (13%)
Length = 668
Score = 72.4 bits (176), Expect = 4e-13
Identities = 41/146 (28%), Positives = 71/146 (48%)
Frame = -1
Query: 232 YASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSW 291
Y +F+ A N+W + Y++ ++SLP I +G+ D+DV + KYP LY G ++
Sbjct: 665 YTAFTLTTAINEWSSTLYSIXYSSLPTIIVGILDKDVGKRTLXKYPQLYGAGQRHVAYNK 486
Query: 292 PRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMAL 351
+ ML+ + S+ IF+ F VD +G VV VN +A+
Sbjct: 485 KLFLLTMLDTLWQSMVIFWA-------PLFAYWSSTVDVASIGDLWTLGVVILVNLHLAM 327
Query: 352 SINYFTWIQHFFIWGSIFFWYVFLLV 377
+ + W+ H IWGSI ++ +++
Sbjct: 326 DVIRWYWVTHAVIWGSIVATFISVMI 249
>BF070523 similar to GP|16930515|gb AT5g44240/MLN1_17 {Arabidopsis thaliana},
partial (24%)
Length = 355
Score = 66.2 bits (160), Expect = 3e-11
Identities = 34/108 (31%), Positives = 59/108 (54%)
Frame = +2
Query: 170 GISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWF 229
GISG EG+QA A ++ + + R+L RL+LVHG + Y R + + Y FY ++ F
Sbjct: 2 GISGREGLQAARADNYIVGKLRYLNRLILVHGRYSYNRTAFLSQYSFYDSLLICFIQNIM 181
Query: 230 EAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYP 277
SG + + NV +T++PV+ V D+D++ + +++P
Sbjct: 182 ALITGVSGTSLSSSVSLMANNVIYTTVPVLE-SVRDRDLNEETVMRHP 322
>BU082774 weakly similar to SP|Q9SX33|ALA9 Potential
phospholipid-transporting ATPase 9 (EC 3.6.3.1)
(Aminophospholipid flippase 9)., partial (6%)
Length = 429
Score = 62.0 bits (149), Expect = 6e-10
Identities = 25/52 (48%), Positives = 36/52 (69%)
Frame = +2
Query: 392 VFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEG 443
VF+E APS +W+ T+ V + L+PYF+Y A Q +F PMYH+++Q R EG
Sbjct: 11 VFIETLAPSPSFWIVTLFVSISTLIPYFSYSAIQMKFFPMYHEMVQWIRHEG 166
>AI855855 similar to SP|Q9SAF5|ALAB Potential phospholipid-transporting
ATPase 11 (EC 3.6.3.1) (Aminophospholipid flippase 11).,
partial (3%)
Length = 148
Score = 60.8 bits (146), Expect = 1e-09
Identities = 27/43 (62%), Positives = 32/43 (73%)
Frame = -1
Query: 325 GQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGS 367
G+ ++L VTMY+ VVW VN QMAL+I YFT IQH FIWGS
Sbjct: 148 GRTAGRDMLAVTMYTCVVWVVNLQMALAIRYFTLIQHIFIWGS 20
>AW279006 weakly similar to SP|P98204|ALA1 Phospholipid-transporting ATPase 1
(EC 3.6.3.1) (Aminophospholipid flippase 1). [Mouse-ear
cress], partial (7%)
Length = 424
Score = 60.1 bits (144), Expect = 2e-09
Identities = 36/124 (29%), Positives = 59/124 (47%)
Frame = +3
Query: 254 TSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTT 313
TS+P I +GV D+D+S K L+YP LY G ++ M++ + SL +F++
Sbjct: 3 TSIPTIVVGVLDKDLSHKTLLQYPKLYGAGHRHEAYNMQLFWFTMIDTLWQSLVLFYI-- 176
Query: 314 NSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYV 373
F +D +G VV VN +A+ IN + + H +WGSI Y
Sbjct: 177 -----PVFIYKDSTIDIWSMGSLWTISVVILVNVHLAMDINQWALVSHVAVWGSIIITYG 341
Query: 374 FLLV 377
+++
Sbjct: 342 CMVI 353
>BF009725 similar to SP|P98204|ALA Phospholipid-transporting ATPase 1 (EC
3.6.3.1) (Aminophospholipid flippase 1). [Mouse-ear
cress], partial (10%)
Length = 400
Score = 50.8 bits (120), Expect = 1e-06
Identities = 22/35 (62%), Positives = 29/35 (82%)
Frame = +2
Query: 5 AGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVI 39
AGIK+WVLTGDK ETA++IGY+ LL +M +I+I
Sbjct: 296 AGIKVWVLTGDKQETAISIGYSSKLLTSNMTQIII 400
>TC221207
Length = 434
Score = 50.1 bits (118), Expect = 2e-06
Identities = 21/51 (41%), Positives = 29/51 (56%)
Frame = +1
Query: 392 VFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVE 442
VF+E APS +W+ T V + L+PY + Q F PMYH ++Q R E
Sbjct: 4 VFIETLAPSPSFWIVTFFVAISTLIPYVSCSVIQMWFFPMYHQMVQWIRYE 156
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,102,129
Number of Sequences: 63676
Number of extensions: 346880
Number of successful extensions: 3183
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 3127
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3167
length of query: 471
length of database: 12,639,632
effective HSP length: 101
effective length of query: 370
effective length of database: 6,208,356
effective search space: 2297091720
effective search space used: 2297091720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0201.1


