

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
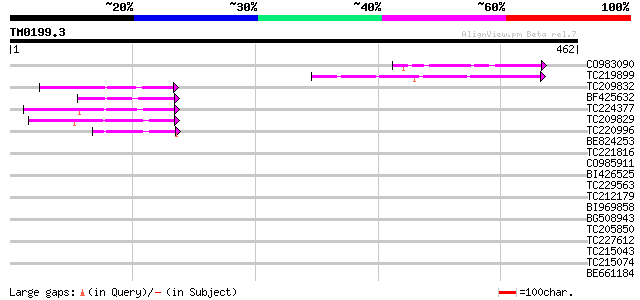
Query= TM0199.3
(462 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO983090 52 4e-07
TC219899 52 6e-07
TC209832 47 2e-05
BF425632 47 2e-05
TC224377 46 4e-05
TC209829 44 1e-04
TC220996 43 3e-04
BE824253 40 0.002
TC221816 weakly similar to UP|Q70G10 (Q70G10) SUMO protease, par... 39 0.006
CO985911 38 0.011
BI426525 37 0.024
TC229563 35 0.071
TC212179 35 0.092
BI969858 homologue to SP|Q9BBS1|ACCD_ Acetyl-coenzyme A carboxyl... 34 0.12
BG508943 34 0.12
TC205850 homologue to UP|Q9LWB3 (Q9LWB3) Ribosomal protein L23, ... 34 0.16
TC227612 weakly similar to UP|Q8L7S0 (Q8L7S0) At1g09730/F21M12_1... 33 0.21
TC215043 similar to UP|Q94KS0 (Q94KS0) Histidine-containing phos... 33 0.35
TC215074 similar to UP|O04237 (O04237) Transcription factor, par... 32 0.46
BE661184 weakly similar to GP|9294523|dbj protein transport prot... 32 0.60
>CO983090
Length = 830
Score = 52.4 bits (124), Expect = 4e-07
Identities = 38/131 (29%), Positives = 65/131 (49%), Gaps = 6/131 (4%)
Frame = -3
Query: 313 FLSPHNL-----SAYESRVSQYIADALKQNEQPNRVTFAPYNMGDHWVLLVIYTSEFAIE 367
FL P ++ S +ES YI ++ ++ R + + HW ++VI E +
Sbjct: 573 FLEPQSI*RSGQSQFESE--SYIKSWMQSSQ---RDVYLGA*LNGHW*MVVILPKEHLVV 409
Query: 368 YFDSFDGEPTDDVHMKNIFDAGLMIYHANSNDVPKKKVKY-IKWRKMKCPKQKNLTDCGY 426
+F S P D ++K I ++ + +D P+ K K +W +KC +QK T+CGY
Sbjct: 408 WFCSLH*RP--DNYLKGIINSAIK----GLDDAPQPKSKASARWIVVKCNRQKGSTECGY 247
Query: 427 YVLKYMRDIII 437
YV+ +M II+
Sbjct: 246 YVMHWMSTIIL 214
>TC219899
Length = 873
Score = 52.0 bits (123), Expect = 6e-07
Identities = 43/194 (22%), Positives = 88/194 (45%), Gaps = 4/194 (2%)
Frame = +2
Query: 247 EDLLKIPMPENILNTNTEEEFVEQIGEEQVNEIYYHTK-LSASVICVYMRFLYSEVLVQN 305
+D +++P ++L+ + F I + E+ ++ L+ SVI +++ +L+S +
Sbjct: 65 KDHVQLPWDPDVLSKPSTMSF--HIYRRDIFELCMGSEMLNISVIQLWLMYLHS-LCTDK 235
Query: 306 QLTERFFFLSPHNLSAYESRVSQ---YIADALKQNEQPNRVTFAPYNMGDHWVLLVIYTS 362
+ F+ P ++ + + ++ Y+ L +E+ + APY DHW LL+I
Sbjct: 236 GNAHIYGFMDPQSIQSVGNNATEVQGYLQSRL--HERKKQCYLAPYLHSDHWQLLIICPK 409
Query: 363 EFAIEYFDSFDGEPTDDVHMKNIFDAGLMIYHANSNDVPKKKVKYIKWRKMKCPKQKNLT 422
+ + S + T + MK I + + Y + + K W +C Q
Sbjct: 410 QNVVVLLCSLH-KKTINKEMKTIVNLAMDEYQRLVGSQSRSRRKKPTWILPRCQTQSKGY 586
Query: 423 DCGYYVLKYMRDII 436
+CGYYV+K M ++
Sbjct: 587 ECGYYVMKQMFTVV 628
>TC209832
Length = 638
Score = 46.6 bits (109), Expect = 2e-05
Identities = 36/114 (31%), Positives = 51/114 (44%), Gaps = 1/114 (0%)
Frame = +2
Query: 25 KADVQPGDSPCYSGEGSCPPSPVILPEG-INACILYLATPTHRKVASGTMYNNSIDTLHS 83
K DV P + + E PS G N C L++ R VA G +Y S T+H+
Sbjct: 41 KPDVGPLATRAITKESCVDPSGNDPDTGDSNKCELHIEEYPPRLVALGRVYKGST-TIHN 217
Query: 84 CPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGTFVAWPKRLVSTM 137
PL D VKV VE + P+P+ + + A+ TF+AW LV +
Sbjct: 218 IPLLHDQVKVGVEEIRDANA----PIPVPTKEVKVMGQALNTFLAWATHLVKRL 367
>BF425632
Length = 334
Score = 46.6 bits (109), Expect = 2e-05
Identities = 27/83 (32%), Positives = 43/83 (51%)
Frame = +2
Query: 56 CILYLATPTHRKVASGTMYNNSIDTLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGN 115
C LY+ R VA G +Y S ++H+ PL D VKV VE + ++ P+P+
Sbjct: 89 CGLYIEEYPPRLVALGRVYEGST-SIHNIPLLHDQVKVGVEDIRD----AHAPIPVPTKE 253
Query: 116 IFTIEDAIGTFVAWPKRLVSTMT 138
+ + A+ TF+AWP L ++
Sbjct: 254 V*VMVQALNTFLAWPTHLAKRLS 322
>TC224377
Length = 454
Score = 45.8 bits (107), Expect = 4e-05
Identities = 37/133 (27%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 12 VHELMKNSGQAQKKADVQPG-DSPC-YSGEGS-CPPSPVILPEGINA---CILYLATPTH 65
V + + + G Q+ +++P +SP S +G+ P P + N C LY+
Sbjct: 37 VRQELASLGLLQQNTNIEPTPNSPINVSTKGNDATPDPSVEDNHTNIRDQCELYVDVDPP 216
Query: 66 RKVASGTMYNNSIDTLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGT 125
VA G +YN T+H + D+V+V VE + E + VP+ + T+ A
Sbjct: 217 HLVAIGKVYNLG-STIHHKTIEDDNVRVVVEEVRDAEAR----VPLPTNEVQTVGQAPNQ 381
Query: 126 FVAWPKRLVSTMT 138
F+ WP+RLV +++
Sbjct: 382 FIQWPRRLVKSIS 420
>TC209829
Length = 530
Score = 44.3 bits (103), Expect = 1e-04
Identities = 34/125 (27%), Positives = 54/125 (43%), Gaps = 2/125 (1%)
Frame = +2
Query: 16 MKNSGQAQKKADVQPGDSPCYSGEGSCPPSPVILPE--GINACILYLATPTHRKVASGTM 73
M++ G A + P S +GSC PE + C LY+ VA G +
Sbjct: 80 MQSQGLAVPPEPLVGPSGPQVSTKGSCADPSGNDPETGDSDRCGLYIEADPAGLVAMGRV 259
Query: 74 YNNSIDTLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGTFVAWPKRL 133
Y +H+ PL VKVSVE + + +P+ + + + TF+AWP L
Sbjct: 260 YEG-FTVVHNTPLLPGQVKVSVEEVTDADAPVLVPI----DEVS*VGQTLHTFLAWPTHL 424
Query: 134 VSTMT 138
V +++
Sbjct: 425 VKSLS 439
>TC220996
Length = 849
Score = 43.1 bits (100), Expect = 3e-04
Identities = 28/79 (35%), Positives = 41/79 (51%), Gaps = 7/79 (8%)
Frame = +1
Query: 68 VASGTMYNNSIDTLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGTFV 127
VA G +Y T+H+ D V+VSVE I+G+ + +P+ I + + TF+
Sbjct: 226 VALGKIYEGG-STIHNMAYADDIVRVSVEKVIDGDAQVSLPM----SEIQYVRQTLHTFI 390
Query: 128 AWPKRLV-------STMTP 139
AWPK LV ST+TP
Sbjct: 391 AWPKPLVKLVSNEDSTITP 447
>BE824253
Length = 567
Score = 40.4 bits (93), Expect = 0.002
Identities = 18/41 (43%), Positives = 27/41 (64%), Gaps = 1/41 (2%)
Frame = -3
Query: 398 NDVPKKKVKY-IKWRKMKCPKQKNLTDCGYYVLKYMRDIII 437
+D P+ K K +W +KC +QK T+CGYYV+ +M II+
Sbjct: 439 DDTPQPKSKAGARWIIVKCNRQKRSTECGYYVMHWMSTIIL 317
>TC221816 weakly similar to UP|Q70G10 (Q70G10) SUMO protease, partial (28%)
Length = 943
Score = 38.5 bits (88), Expect = 0.006
Identities = 28/90 (31%), Positives = 41/90 (45%), Gaps = 3/90 (3%)
Frame = +3
Query: 345 FAPYNMGDHWVLLVIYTSEFAIEYFDSFDGEPTDDVHMKNIFDAGLMIYHANSNDVPKKK 404
F P + HW L VI + +Y DS G TD MK + A ++ D K
Sbjct: 306 FVPIHKEIHWCLAVINKKDKKFQYLDSLRG--TDAQVMKIL--ASYIVDEV--KDKTGKD 467
Query: 405 VKYIKWRK---MKCPKQKNLTDCGYYVLKY 431
+ W+K P+Q+N DCG +++KY
Sbjct: 468 IDVSSWKKEFVEDLPEQQNGYDCGVFMIKY 557
>CO985911
Length = 825
Score = 37.7 bits (86), Expect = 0.011
Identities = 21/55 (38%), Positives = 33/55 (59%)
Frame = +2
Query: 80 TLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGTFVAWPKRLV 134
T+H+ PL +D VKV VE + + ++IPVP + + + A+ F+AWP LV
Sbjct: 470 TVHNIPLDNDLVKVGVEEVQDVD--AHIPVPTQ--KVQLVGQALNIFLAWPTHLV 622
>BI426525
Length = 422
Score = 36.6 bits (83), Expect = 0.024
Identities = 21/71 (29%), Positives = 35/71 (48%)
Frame = +3
Query: 68 VASGTMYNNSIDTLHSCPLPSDHVKVSVEFAIEGEEKSYIPVPIEGGNIFTIEDAIGTFV 127
VA G +Y +H+ PL VKVSVE + + +P+ + + + TF+
Sbjct: 42 VAMGRVYEG-FTVVHNTPLLPGQVKVSVEEVTDADAPVLVPI----DEVS*VGQTLHTFL 206
Query: 128 AWPKRLVSTMT 138
AWP LV +++
Sbjct: 207 AWPTHLVKSLS 239
>TC229563
Length = 669
Score = 35.0 bits (79), Expect = 0.071
Identities = 30/121 (24%), Positives = 55/121 (44%), Gaps = 1/121 (0%)
Frame = +1
Query: 143 DNNHKATSRAKGITAVQAKKPAGQVKKPAERANKPVAQGEKVHEQAKKQVDKGKKDVGQG 202
D ++ T + I +P + +KPAE KP + EK E+AKK+ K+ G
Sbjct: 43 DYVYRRTKKQAKIVPQPEPEPEKKEEKPAEEETKP--EEEKPPEEAKKEEGGENKEEKGG 216
Query: 203 KRAPGQAKKQADKGTEIQPLGASSKDKLQNCKFVWAY-ARSVLKSEDLLKIPMPENILNT 261
+ + +AKK+ + + P + L+ + + Y SV++ +IP P+ +
Sbjct: 217 EESKEEAKKE-ENNVVVVPYNNIDDEGLKRMMYYYQYPPLSVIE-----RIPPPQLFSDE 378
Query: 262 N 262
N
Sbjct: 379 N 381
>TC212179
Length = 764
Score = 34.7 bits (78), Expect = 0.092
Identities = 19/43 (44%), Positives = 25/43 (57%)
Frame = +1
Query: 415 CPKQKNLTDCGYYVLKYMRDIIIAGDSQTLQEVVILVIKYIVF 457
C +QK T+CGYYV+ M II+ G + E VI+ K I F
Sbjct: 391 CNRQKGSTECGYYVMHCMSTIIL-GTFKNNWETVIV*FKQISF 516
>BI969858 homologue to SP|Q9BBS1|ACCD_ Acetyl-coenzyme A carboxylase carboxyl
transferase subunit beta (EC 6.4.1.2) (ACCASE beta
chain)., partial (2%)
Length = 694
Score = 34.3 bits (77), Expect = 0.12
Identities = 12/23 (52%), Positives = 18/23 (78%)
Frame = -1
Query: 415 CPKQKNLTDCGYYVLKYMRDIII 437
C +QK +T+CGYYV+ +M II+
Sbjct: 295 CNRQKGITECGYYVMHWMSTIIL 227
>BG508943
Length = 383
Score = 34.3 bits (77), Expect = 0.12
Identities = 13/23 (56%), Positives = 17/23 (73%)
Frame = -1
Query: 415 CPKQKNLTDCGYYVLKYMRDIII 437
C KQK T+CGYYV+ +M II+
Sbjct: 98 CNKQKGCTECGYYVMHWMSTIIL 30
>TC205850 homologue to UP|Q9LWB3 (Q9LWB3) Ribosomal protein L23, partial
(94%)
Length = 679
Score = 33.9 bits (76), Expect = 0.16
Identities = 20/48 (41%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = +1
Query: 132 RLVSTMTPASSDNNHKATSRAKGITAVQAKKPAGQV-KKPAERANKPV 178
R V TM PA +DN KA +A+ + +A K GQV KK A++ V
Sbjct: 43 R*VITMAPAKADNAKKADPKAQALKTAKAVKSGGQVFKKKAKKIRTSV 186
>TC227612 weakly similar to UP|Q8L7S0 (Q8L7S0) At1g09730/F21M12_12, partial
(19%)
Length = 1563
Score = 33.5 bits (75), Expect = 0.21
Identities = 16/55 (29%), Positives = 32/55 (58%), Gaps = 4/55 (7%)
Frame = +1
Query: 382 MKNIFDAGLMIY----HANSNDVPKKKVKYIKWRKMKCPKQKNLTDCGYYVLKYM 432
+KN+F + L H+N + K ++++ ++ P+Q+NL DCG ++L Y+
Sbjct: 130 LKNVFQSYLCEEWKERHSNVVEDVSSKFLHLRFISLELPQQENLYDCGLFLLHYV 294
>TC215043 similar to UP|Q94KS0 (Q94KS0) Histidine-containing phosphotransfer
protein, partial (98%)
Length = 923
Score = 32.7 bits (73), Expect = 0.35
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Frame = +2
Query: 11 LVHELMKNSGQAQKKADVQPGDSPCYSGEGSCPPS-PVILPEGINACILYLATPTHRKVA 69
+VHE S ++ + PG SP + E + P+ +ILP + C+ T +
Sbjct: 29 MVHEK*SKSQSQRRIKSLSPGASPVHCREQTPTPNLKIILPLCFSLCLSQKTTQKNHGRG 208
Query: 70 SGTMYNNSIDTLHSCPLP 87
S ++D LH PLP
Sbjct: 209 SDA---ETVDRLHQIPLP 253
>TC215074 similar to UP|O04237 (O04237) Transcription factor, partial (88%)
Length = 1720
Score = 32.3 bits (72), Expect = 0.46
Identities = 20/64 (31%), Positives = 31/64 (48%)
Frame = +1
Query: 147 KATSRAKGITAVQAKKPAGQVKKPAERANKPVAQGEKVHEQAKKQVDKGKKDVGQGKRAP 206
KA + A TA K PA Q+K PA+ +KP+ + V + + V G+ G R
Sbjct: 322 KAAAAAAAATAAPKKAPAQQIK-PAQLPSKPLPPAQAVRDARNESVRGGRGGGRGGGRGF 498
Query: 207 GQAK 210
G+ +
Sbjct: 499 GRGR 510
>BE661184 weakly similar to GP|9294523|dbj protein transport protein Sec23
{Arabidopsis thaliana}, partial (20%)
Length = 645
Score = 32.0 bits (71), Expect = 0.60
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = -3
Query: 35 CYSGEGSCPPSPVILPEGINACILYLATPTHRKVASG 71
C G+GS PPSP PEG C++ ++ R +SG
Sbjct: 274 CQGGDGSEPPSP---PEGSRTCLIPPSSAATRSTSSG 173
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,130,416
Number of Sequences: 63676
Number of extensions: 259406
Number of successful extensions: 1483
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1470
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1479
length of query: 462
length of database: 12,639,632
effective HSP length: 101
effective length of query: 361
effective length of database: 6,208,356
effective search space: 2241216516
effective search space used: 2241216516
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0199.3


