

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0197.2
(366 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
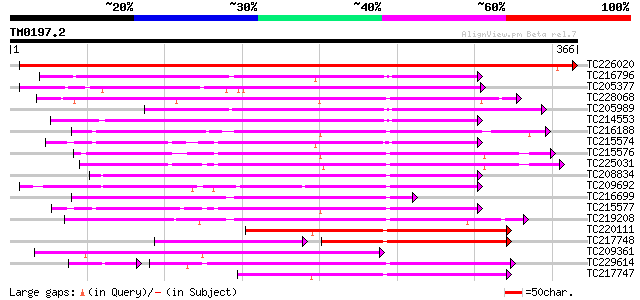
Score E
Sequences producing significant alignments: (bits) Value
TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, ... 651 0.0
TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {A... 202 1e-52
TC205377 186 2e-47
TC228068 anthocyanidin synthase [Glycine max] 175 3e-44
TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-l... 173 1e-43
TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 168 4e-42
TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-car... 165 3e-41
TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%) 163 1e-40
TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%) 159 1e-39
TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-c... 157 5e-39
TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 155 4e-38
TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1... 149 2e-36
TC216699 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial... 146 1e-35
TC215577 homologue to UP|Q41681 (Q41681) 1-aminocylopropane-1-ca... 146 2e-35
TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, part... 144 8e-35
TC220111 similar to PIR|T49209|T49209 leucoanthocyanidin dioxyge... 140 7e-34
TC217748 121 9e-34
TC209361 weakly similar to UP|Q7XJ89 (Q7XJ89) Flavonol synthase ... 138 4e-33
TC229614 similar to UP|O48631 (O48631) Ethylene-forming-enzyme-l... 128 6e-33
TC217747 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyg... 137 8e-33
>TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(93%)
Length = 1441
Score = 651 bits (1679), Expect = 0.0
Identities = 322/370 (87%), Positives = 339/370 (91%), Gaps = 10/370 (2%)
Frame = +3
Query: 7 KTLTTLAQQNTLESSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIVEA 66
KTLT LAQ+ TLESSFVRDE+ERPKVAYN FS+EIPVISLAGIDEVDGRR EIC KIVEA
Sbjct: 78 KTLTYLAQEKTLESSFVRDEEERPKVAYNEFSDEIPVISLAGIDEVDGRRREICEKIVEA 257
Query: 67 CENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGES 126
CENWGIFQVVDHGVD +LV+ MT LAKEFFALPP+EKLRFDM+G KKGGFIVSSHLQGES
Sbjct: 258 CENWGIFQVVDHGVDQQLVAEMTRLAKEFFALPPDEKLRFDMSGAKKGGFIVSSHLQGES 437
Query: 127 VQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEK 186
VQDWREIVTYFSYP R RDYSRWPDTP GW++VTEEYS+K+MGLACKL+EVLSEAMGLEK
Sbjct: 438 VQDWREIVTYFSYPKRERDYSRWPDTPEGWRSVTEEYSDKVMGLACKLMEVLSEAMGLEK 617
Query: 187 EALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKT 246
E L+KACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKT
Sbjct: 618 EGLSKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKT 797
Query: 247 WITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPL 306
WITVQPVE AFVVNLGDH HYLSNGRFKNADHQAVVNSN SRLSIATFQNPAP+ATVYPL
Sbjct: 798 WITVQPVEAAFVVNLGDHAHYLSNGRFKNADHQAVVNSNHSRLSIATFQNPAPNATVYPL 977
Query: 307 KVREGEKSVMEEPITFAEMYRRKMSKDIELARMKKLAKEKKLQDLE----------KAKL 356
K+REGEK VMEEPITFAEMYRRKMSKDIE+ARMKKLAKEK LQDLE KAKL
Sbjct: 978 KIREGEKPVMEEPITFAEMYRRKMSKDIEIARMKKLAKEKHLQDLENEKHLQELDQKAKL 1157
Query: 357 EPKPMNEIFA 366
E KP+ EI A
Sbjct: 1158EAKPLKEILA 1187
>TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {Arabidopsis
thaliana;} , partial (72%)
Length = 1454
Score = 202 bits (515), Expect = 1e-52
Identities = 113/293 (38%), Positives = 172/293 (58%), Gaps = 7/293 (2%)
Frame = +1
Query: 20 SSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDG-RRSEICNKIVEACENWGIFQVVDH 78
S+F+R +RP + S+++ I L + ++ G RS I +I +AC+N+G FQV +H
Sbjct: 145 SNFIRPLGDRPNLQGVVQSSDV-CIPLIDLQDLHGPNRSHIIQQIDQACQNYGFFQVTNH 321
Query: 79 GVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSS-HLQGESVQDWREIVTYF 137
GV ++ + + +EFF LP EKL+ T K + +S ++ E V WR+ +
Sbjct: 322 GVPEGVIEKIMKVTREFFGLPESEKLKSYSTDPFKASRLSTSFNVNSEKVSSWRDFLRLH 501
Query: 138 SYPIRNRDYSR-WPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDM 196
+PI DY + WP P + EY K+ G++ KL+E +SE++GLE++ + +
Sbjct: 502 CHPIE--DYIKEWPSNPPSLREDVAEYCRKMRGVSLKLVEAISESLGLERDYINRVVGGK 675
Query: 197 ----DQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQP 252
Q + +NYYP CP+P+LT GL HTDP IT+LLQD+V GLQ +D GK W+ V P
Sbjct: 676 KGQEQQHLAMNYYPACPEPELTYGLPGHTDPTVITILLQDEVPGLQVLKD-GK-WVAVNP 849
Query: 253 VEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 305
+ FVVN+GD +SN ++K+ H+AVVN N R+SI TF P+ DA + P
Sbjct: 850 IPNTFVVNVGDQIQVISNDKYKSVLHRAVVNCNKDRISIPTFYFPSNDAIIGP 1008
>TC205377
Length = 1345
Score = 186 bits (471), Expect = 2e-47
Identities = 106/321 (33%), Positives = 178/321 (55%), Gaps = 20/321 (6%)
Frame = +3
Query: 7 KTLTTLAQQNTLESSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDGRRSE------IC 60
K LT ++++F++D RPK++ + IP+I L+ I + R S+ +
Sbjct: 66 KLLTQSPAMGEVDAAFIQDPPHRPKLSTIQ-AEGIPIIDLSPI--TNHRVSDPSAIEGLV 236
Query: 61 NKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSS 120
+I AC WG FQV +HGV L ++ +K FFA EEK + G+ +
Sbjct: 237 KEIGSACNEWGFFQVTNHGVPLTLRQNIEKASKLFFAQSAEEKRKVSRNESSPAGYYDTE 416
Query: 121 HLQGESVQDWREIVTYFS-----YPIRNRDY----SRW----PDTPAGWKAVTEEYSEKL 167
H + +V+DW+E+ + + P+ + ++ ++W P+ P ++ VT+EY +++
Sbjct: 417 HTK--NVRDWKEVFDFLAKEPTFIPVTSDEHDDRVNQWTNQSPEYPLNFRVVTQEYIQEM 590
Query: 168 MGLACKLLEVLSEAMGLEKEALTKACV-DMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTI 226
L+ K+LE+++ ++GLE + + + + D + +N+YP CP PDL LG+ RH DPG +
Sbjct: 591 EKLSFKILELIALSLGLEAKRVEEFFIKDQTSFIRLNHYPPCPYPDLALGVGRHKDPGAL 770
Query: 227 TLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNS 286
T+L QD+VGGL+ R + WI V+P A+++N+GD SN +++ DH+ VVNS
Sbjct: 771 TILAQDEVGGLEVRRKADQEWIRVKPTPDAYIINIGDTVQVWSNDAYESVDHRVVVNSEK 950
Query: 287 SRLSIATFQNPAPDATVYPLK 307
R SI F PA D V PL+
Sbjct: 951 ERFSIPFFFFPAHDTEVKPLE 1013
>TC228068 anthocyanidin synthase [Glycine max]
Length = 1231
Score = 175 bits (444), Expect = 3e-44
Identities = 109/328 (33%), Positives = 169/328 (51%), Gaps = 15/328 (4%)
Frame = +3
Query: 18 LESSFVRDEDERPKVAYNNFSNE------IPVISLAGIDEVDGR-RSEICNKIVEACENW 70
+ +VR ++E + N F E +P I L ID D R + K+ +A E W
Sbjct: 57 IPKEYVRPQEELKSIG-NVFEEEKKEGLQVPTIDLREIDSEDEVVRGKCREKLKKAAEEW 233
Query: 71 GIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRF--DMTGGKKGGFIVSSHLQGESVQ 128
G+ +V+HG+ EL+ + + FF L EEK ++ D+ GK G+
Sbjct: 234 GVMHLVNHGIPDELIERVKKAGETFFGLAVEEKEKYANDLESGKIQGYGSKLANNASGQL 413
Query: 129 DWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEA 188
+W + + ++P RD S WP PA + VT EY+++L GLA K+LE LS +GLE
Sbjct: 414 EWEDYFFHLAFPEDKRDLSFWPKKPADYIEVTSEYAKRLRGLATKILEALSIGLGLEGRR 593
Query: 189 LTKACVDMDQ---KVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGK 245
L K M++ ++ +NYYP CPQP+L LG++ HTD ++T LL + V GLQ
Sbjct: 594 LEKEVGGMEELLLQLKINYYPICPQPELALGVEAHTDVSSLTFLLHNMVPGLQLFYQG-- 767
Query: 246 TWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATV-- 303
W+T + V + ++++GD LSNG++K+ H+ +VN R+S A F P + +
Sbjct: 768 QWVTAKCVPNSILMHIGDTIEILSNGKYKSILHRGLVNKEKVRISWAVFCEPPKEKIILQ 947
Query: 304 -YPLKVREGEKSVMEEPITFAEMYRRKM 330
P V E E + P TFA+ K+
Sbjct: 948 PLPELVTETEPARF-PPRTFAQHIHHKL 1028
>TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-like protein,
partial (74%)
Length = 1192
Score = 173 bits (439), Expect = 1e-43
Identities = 99/262 (37%), Positives = 148/262 (55%), Gaps = 3/262 (1%)
Frame = +2
Query: 88 MTTLAKEFFALPPEEKLR-FDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDY 146
M +A FF LP EEKL+ + K S +++ E+V++WR+ + YP+ +
Sbjct: 11 MEEVAHGFFKLPVEEKLKLYSEDTSKTMRLSTSFNVKKETVRNWRDYLRLHCYPLE-KYA 187
Query: 147 SRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYYP 206
WP P +K EY + L ++ E +SE++GLEK+ + + Q + VNYYP
Sbjct: 188 PEWPSNPPSFKETVTEYCTIIRELGLRIQEYISESLGLEKDYIKNVLGEQGQHMAVNYYP 367
Query: 207 KCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGDHG 265
CP+P+LT GL HTDP +T+LLQD QV GLQ +D GK W+ V P AFV+N+GD
Sbjct: 368 PCPEPELTYGLPGHTDPNALTILLQDLQVAGLQVLKD-GK-WLAVSPQPNAFVINIGDQL 541
Query: 266 HYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK-VREGEKSVMEEPITFAE 324
LSNG +K+ H+AVVN RLS+A+F P +A + P K + E + T+AE
Sbjct: 542 QALSNGLYKSVWHRAVVNVEKPRLSVASFLCPNDEALISPAKPLTEHGSEAVYRGFTYAE 721
Query: 325 MYRRKMSKDIELARMKKLAKEK 346
Y++ S++++ +L K K
Sbjct: 722 YYKKFWSRNLDQEHCLELFKNK 787
>TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (52%)
Length = 1808
Score = 168 bits (425), Expect = 4e-42
Identities = 93/281 (33%), Positives = 156/281 (55%), Gaps = 2/281 (0%)
Frame = +1
Query: 27 DERPKVAYNNFS-NEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELV 85
++ P + N S ++P+I L + D E K+ AC+ WG FQ+++HGVD +V
Sbjct: 121 NQDPHILSNTISLPQVPIIDLHQLLSEDPSELE---KLDHACKEWGFFQLINHGVDPPVV 291
Query: 86 SHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRD 145
+M +EFF LP EEK +F T GF + E +W ++ ++P+ +R+
Sbjct: 292 ENMKIGVQEFFNLPMEEKQKFWQTPEDMQGFGQLFVVSEEQKLEWADMFYAHTFPLHSRN 471
Query: 146 YSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYY 205
P P ++ E Y +L + ++ ++ +A+ ++ L++ D Q + +NYY
Sbjct: 472 PHLIPKIPQPFRENLENYCLELRKMCITIIGLMKKALKIKTNELSELFEDPSQGIRMNYY 651
Query: 206 PKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQATRDNGKTWITVQPVEGAFVVNLGDH 264
P CPQP+ +G+ H+D G +T+LLQ ++V GLQ +D GK WI V+P+ AFV+N+GD
Sbjct: 652 PPCPQPERVIGINPHSDSGALTILLQVNEVEGLQIRKD-GK-WIPVKPLSNAFVINVGDM 825
Query: 265 GHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 305
L+NG +++ +H+ +VNS R+SIA F P + P
Sbjct: 826 LEILTNGIYRSIEHRGIVNSEKERISIAMFHRPQMSRVIGP 948
>TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-carboxylic acid
oxidase, complete
Length = 1362
Score = 165 bits (418), Expect = 3e-41
Identities = 106/317 (33%), Positives = 163/317 (50%), Gaps = 8/317 (2%)
Frame = +3
Query: 41 IPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPP 100
+PVI + ++ + R++ +I CE WG FQ+++HG+ EL+ + +A EF+ L
Sbjct: 147 VPVIDFSKLNGEE--RTKTMAQIANGCEEWGFFQLINHGIPEELLERVKKVASEFYKLER 320
Query: 101 EEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVT 160
EE + + + + E V DW +++T D + WP+ G++
Sbjct: 321 EENFKNSTSVKLLSDSVEKKSSEMEHV-DWEDVITLL-------DDNEWPEKTPGFRETM 476
Query: 161 EEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQK-----VVVNYYPKCPQPDLTL 215
EY +L LA KL+EV+ E +GL K + KA D + V++YP CP P+L
Sbjct: 477 AEYRAELKKLAEKLMEVMDENLGLTKGYIKKALNGGDGENAFFGTKVSHYPPCPHPELVK 656
Query: 216 GLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFK 274
GL+ HTD G + LL QD +VGGLQ ++ WI VQP+ A V+N GD LSNGR+K
Sbjct: 657 GLRAHTDAGGVILLFQDDKVGGLQMLKEG--QWIDVQPLPNAIVINTGDQIEVLSNGRYK 830
Query: 275 NADHQAVVNSNSSRLSIATFQNPAPDATVYPLKVREGEKSVMEEPITFAEMYRRKMSKDI 334
+ H+ + + +R SIA+F NP+ AT+ P + V +E E Y + + D
Sbjct: 831 SCWHRVLATPDGNRRSIASFYNPSFKATICP-----APQLVEKEDQQVDETYPKFVFGDY 995
Query: 335 --ELARMKKLAKEKKLQ 349
A K L KE + Q
Sbjct: 996 MSVYAEQKFLPKEPRFQ 1046
>TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%)
Length = 1169
Score = 163 bits (412), Expect = 1e-40
Identities = 107/288 (37%), Positives = 160/288 (55%), Gaps = 6/288 (2%)
Frame = +3
Query: 24 RDEDERPKV-AYNNFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDT 82
R E E+ K NF PVI+L ++ + R N+I +AC+NWG F++V+HG+
Sbjct: 6 RREKEKEKA*EMENF----PVINLENLNGEE--RKATLNQIEDACQNWGFFELVNHGIPL 167
Query: 83 ELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGE-SVQDWREIVTYFSYPI 141
EL+ + L KE + E+ RF KG L+ E DW T+F +
Sbjct: 168 ELLDTVERLTKEHYRKCMEK--RFKEAVSSKG-------LEAEVKDMDWES--TFFLRHL 314
Query: 142 RNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDM---DQ 198
+ S PD ++ +E+++KL LA +LL++L E +GLEK L A +
Sbjct: 315 PTSNISEIPDLSQEYRDAMKEFAQKLEKLAEELLDLLCENLGLEKGYLKNAFYGSRGPNF 494
Query: 199 KVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAF 257
V YP CP+P+L GL+ HTD G I LLLQD +V GLQ + NG+ W+ V P+ +
Sbjct: 495 GTKVANYPACPKPELVKGLRAHTDAGGIILLLQDDKVSGLQLLK-NGQ-WVDVPPMRHSI 668
Query: 258 VVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 305
VVNLGD ++NGR+K+ +H+ + +N +R+S+A+F NPA DA +YP
Sbjct: 669 VVNLGDQIEVITNGRYKSVEHRVIAQTNGTRMSVASFYNPASDALIYP 812
>TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%)
Length = 1151
Score = 159 bits (403), Expect = 1e-39
Identities = 108/320 (33%), Positives = 170/320 (52%), Gaps = 9/320 (2%)
Frame = +2
Query: 42 PVISLAGIDEVDGR-RSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPP 100
PVI+L D ++G R ++I +AC+NWG F++V+HG+ EL+ + L KE +
Sbjct: 2 PVINL---DNLNGEERKATLDQIEDACQNWGFFELVNHGIPLELLDTVERLTKEHYRKCM 172
Query: 101 EEKLRFDMTGGKKGGFIVSSHLQGE-SVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAV 159
E + + + S L+ E DW T+F + + S PD ++
Sbjct: 173 ENRFK---------EAVASKALEVEVKDMDWES--TFFLRHLPTSNISEIPDLSQEYRDA 319
Query: 160 TEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQK---VVVNYYPKCPQPDLTLG 216
+E+++KL LA +LL++L E +GLEK L A V YP CP+P+L G
Sbjct: 320 MKEFAKKLEKLAEELLDLLCENLGLEKGYLKNAFYGSKGPNFGTKVANYPACPKPELVKG 499
Query: 217 LKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKN 275
L+ HTD G I LLLQD +V GLQ +D+ W+ V P+ + VVNLGD ++NGR+K+
Sbjct: 500 LRAHTDAGGIILLLQDDKVSGLQLLKDD--QWVDVPPMRHSIVVNLGDQIEVITNGRYKS 673
Query: 276 ADHQAVVNSNSSRLSIATFQNPAPDATVYP---LKVREGEKSVMEEPITFAEMYRRKMSK 332
+H+ V ++ +R+S+A+F NPA DA +YP L +E +++ P E Y +
Sbjct: 674 VEHRVVARTDGTRMSVASFYNPANDAVIYPAPALLEKEAQETEQVYPKFVFEDYMKL--- 844
Query: 333 DIELARMKKLAKEKKLQDLE 352
A +K KE + Q ++
Sbjct: 845 ---YATLKFQPKEPRFQAIK 895
>TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1244
Score = 157 bits (398), Expect = 5e-39
Identities = 104/321 (32%), Positives = 168/321 (51%), Gaps = 8/321 (2%)
Frame = +1
Query: 46 LAGIDEVDGR-RSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKL 104
L ++++ G R++ KI +ACENWG F++V+HG+ +++ + L KE + EE
Sbjct: 127 LINLEKLSGEERNDTMEKIKDACENWGFFELVNHGIPHDILDTVERLTKEHYRKCMEE-- 300
Query: 105 RFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYS 164
RF KG V + ++ DW T+ + + S PD ++ V ++++
Sbjct: 301 RFKELVASKGLDAVQTEVKD---MDWES--TFHLRHLPESNISEIPDLIDEYRKVMKDFA 465
Query: 165 EKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVV---VNYYPKCPQPDLTLGLKRHT 221
+L LA +LL++L E +GLEK L KA V YP CP P+L GL+ HT
Sbjct: 466 LRLEKLAEQLLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPPCPNPELVKGLRPHT 645
Query: 222 DPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQA 280
D G I LL QD +V GLQ +D W+ V P+ + VVN+GD ++NG++K+ +H+
Sbjct: 646 DAGGIILLFQDDKVSGLQLLKDG--QWVDVPPMRHSIVVNIGDQLEVITNGKYKSVEHRV 819
Query: 281 VVNSNSSRLSIATFQNPAPDATVYP---LKVREGEKSVMEEPITFAEMYRRKMSKDIELA 337
+ ++ +R+SIA+F NP DA +YP L +E E+ P E Y + A
Sbjct: 820 IAQTDGTRMSIASFYNPGSDAVIYPAPELLEKEAEEKNQLYPKFVFEDYMKL------YA 981
Query: 338 RMKKLAKEKKLQDLEKAKLEP 358
++K AKE + + + + P
Sbjct: 982 KLKFQAKEPRFEAFKASNFGP 1044
>TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (54%)
Length = 1070
Score = 155 bits (391), Expect = 4e-38
Identities = 84/255 (32%), Positives = 145/255 (55%), Gaps = 1/255 (0%)
Frame = +2
Query: 52 VDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGG 111
V+ SE+ +K+ AC+ WG FQ+V+HGV++ LV + ++FF LP EK +F T
Sbjct: 5 VESGSSEL-DKLHLACKEWGFFQLVNHGVNSSLVEKVRLETQDFFNLPMSEKKKFWQTPQ 181
Query: 112 KKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLA 171
GF + + + DW ++ + P +R +P P ++ E YS ++ LA
Sbjct: 182 HMEGFGQAFVVSEDQKLDWADLYYMTTLPKHSRMPHLFPQLPLPFRDTLEAYSREIKDLA 361
Query: 172 CKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ 231
++ ++ +A+ +++ + + D Q + +NYYP CP+P+ +GL H+D + +LLQ
Sbjct: 362 IVIIGLMGKALKIQEREIRELFEDGIQLMRMNYYPPCPEPEKVIGLTPHSDGIGLAILLQ 541
Query: 232 -DQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLS 290
++V GLQ +D W+ V+P+ AF+VN+GD ++NG +++ +H+A VN RLS
Sbjct: 542 LNEVEGLQIRKDG--LWVPVKPLINAFIVNVGDILEIITNGIYRSIEHRATVNGEKERLS 715
Query: 291 IATFQNPAPDATVYP 305
ATF +P+ D V P
Sbjct: 716 FATFYSPSSDGVVGP 760
>TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (36%)
Length = 1229
Score = 149 bits (376), Expect = 2e-36
Identities = 98/305 (32%), Positives = 162/305 (52%), Gaps = 6/305 (1%)
Frame = +2
Query: 7 KTLTTLAQQNTLESSFVRDEDERPKV-AYNNFSNEIPVISLAGIDEVDGRRSEICNKIVE 65
K LT++ Q+ +++ + P + A FS+ +P I+L + + E+ K+
Sbjct: 50 KPLTSVPQR------YIQLHNNEPSLLAGETFSHALPTINLKKLIHGEDIELEL-EKLTS 208
Query: 66 ACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGF--IVSSHLQ 123
AC +WG FQ+V+HG+ + ++ + + FF LP EEK+++ + G G+ ++ S
Sbjct: 209 ACRDWGFFQLVEHGISSVVMKTLEDEVEGFFMLPMEEKMKYKVRPGDVEGYGTVIGSE-- 382
Query: 124 GESVQDW--REIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEA 181
+ DW R + IRN +P+ P+ + + E Y E+L LA L+ +L +
Sbjct: 383 -DQKLDWGDRLFMKINXRSIRNPHL--FPELPSSLRNILELYIEELQNLAMILMGLLGKT 553
Query: 182 MGLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQAT 240
+ +EK L + D Q + + YYP CPQP+L +GL H+D IT+L Q + V GLQ
Sbjct: 554 LKIEKREL-EVFEDGIQNMRMTYYPPCPQPELVMGLSAHSDATGITILNQMNGVNGLQIK 730
Query: 241 RDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPD 300
+D WI V + A VVN+GD +SNG +K+ +H+A VNS R+S+A F P
Sbjct: 731 KDG--VWIPVNVISEALVVNIGDIIEIMSNGAYKSVEHRATVNSEKERISVAMFFLPKFQ 904
Query: 301 ATVYP 305
+ + P
Sbjct: 905 SEIGP 919
>TC216699 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (53%)
Length = 1436
Score = 146 bits (369), Expect = 1e-35
Identities = 82/224 (36%), Positives = 123/224 (54%), Gaps = 1/224 (0%)
Frame = +1
Query: 41 IPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPP 100
IP+I L GI + R + K+ ACE WG FQV++HG+ T ++ M F
Sbjct: 439 IPIIDLTGIHDDPILRDHVVGKVRYACEKWGFFQVINHGIPTHVLDEMIKGTCRFHQQDA 618
Query: 101 E-EKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAV 159
+ K + +K ++ + L + DWR+ + + P + PA + +
Sbjct: 619 KVRKEYYTREVSRKVAYLSNYTLFEDPSADWRDTLAFSLAP----HPPEAEEFPAVCRDI 786
Query: 160 TEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKR 219
EYS+K+M LA L E+LSEA+GL + L + Q ++ +YYP CP+P+LT+G +
Sbjct: 787 VNEYSKKIMALAYALFELLSEALGLNRFYLKEMDCAEGQLLLCHYYPACPEPELTMGNTK 966
Query: 220 HTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGD 263
H+D TIT+LLQDQ+GGLQ D+ WI V P+ GA VVN+GD
Sbjct: 967 HSDGNTITILLQDQIGGLQVLHDS--QWIDVPPMHGALVVNIGD 1092
>TC215577 homologue to UP|Q41681 (Q41681) 1-aminocylopropane-1-carboxylate
oxidase homolog , complete
Length = 1382
Score = 146 bits (368), Expect = 2e-35
Identities = 96/282 (34%), Positives = 149/282 (52%), Gaps = 4/282 (1%)
Frame = +2
Query: 28 ERPKVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSH 87
ER K NF PV+ + ++ + R I +ACENWG F++V+HG+ EL+
Sbjct: 32 ERRKKQMANF----PVVDMGKLNTEE--RPAAMEIIKDACENWGFFELVNHGISIELMDT 193
Query: 88 MTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYS 147
+ L KE + E++ + +T KG V S + DW T+F + + S
Sbjct: 194 VEKLTKEHYKKTMEQRFKEMVTS--KGLESVQSEIND---LDWES--TFFLRHLPLSNVS 352
Query: 148 RWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQK---VVVNY 204
D ++ ++++ +L LA +LL++L E +GLEK L K V+
Sbjct: 353 DNADLDQDYRKTMKKFALELEKLAEQLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSN 532
Query: 205 YPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGD 263
YP CP PDL GL+ HTD G I LL QD +V GLQ +D+ WI V P+ + V+NLGD
Sbjct: 533 YPPCPTPDLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDD--QWIDVPPMRHSIVINLGD 706
Query: 264 HGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 305
++NG++K+ H+ + ++ +R+SIA+F NP DA + P
Sbjct: 707 QLEVITNGKYKSVMHRVIAQADDTRMSIASFYNPGDDAVISP 832
>TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, partial (87%)
Length = 1242
Score = 144 bits (362), Expect = 8e-35
Identities = 99/308 (32%), Positives = 145/308 (46%), Gaps = 8/308 (2%)
Frame = +1
Query: 36 NFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEF 95
N IPVI LA + + R EI +I EA E WG FQVV+HG+ ++ + + F
Sbjct: 205 NTEYTIPVIDLAEVGKDPSSRQEIIGRIREASERWGFFQVVNHGIPVTVLEDLKDGVQRF 384
Query: 96 FALPPEEKLRFDMTGGKKGGFIVSSH--LQGESVQDWREIVTYFSYPIRNRDYSRWPDTP 153
EEK T + +S+ L +WR+ + P + + D P
Sbjct: 385 HEQDIEEKKEL-YTRDHMKPLVYNSNFDLYSSPALNWRDSFMCYLAP----NPPKPEDLP 549
Query: 154 AGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYYPKCPQPDL 213
+ + EY +M L L E+LSEA+GL + L + +YYP CP+PDL
Sbjct: 550 VVCRDILLEYGTYVMKLGIALFELLSEALGLHPDHLKDLGCAEGLISLCHYYPACPEPDL 729
Query: 214 TLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRF 273
TLG +H+D +T+LLQD +GGLQ N WI + P GA VVN+GD ++N RF
Sbjct: 730 TLGTTKHSDNCFLTVLLQDHIGGLQVLYQN--MWIDITPEPGALVVNIGDLLQLITNDRF 903
Query: 274 KNADHQAVVNSNSSRLSIATF------QNPAPDATVYPLKVREGEKSVMEEPITFAEMYR 327
K+ +H+ N R+S+A F +P P + L + E T AE R
Sbjct: 904 KSVEHRVQANLIGPRISVACFFSEGLKSSPKPYGPIKELLTEDNPPKYRE--TTVAEYVR 1077
Query: 328 RKMSKDIE 335
+K ++
Sbjct: 1078YFEAKGLD 1101
>TC220111 similar to PIR|T49209|T49209 leucoanthocyanidin dioxygenase-like
protein - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (43%)
Length = 786
Score = 140 bits (354), Expect = 7e-34
Identities = 75/177 (42%), Positives = 112/177 (62%), Gaps = 5/177 (2%)
Frame = +3
Query: 153 PAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACV---DMDQKVVVNYYPKCP 209
P + + EY E+++ L ++LE+LS +GL ++ L A D+ + VN+YPKCP
Sbjct: 15 PTSLRNIISEYGEQVVMLGGRILEILSINLGLREDFLLNAFGGENDLGACLRVNFYPKCP 194
Query: 210 QPDLTLGLKRHTDPGTITLLLQDQ-VGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYL 268
QPDLTLGL H+DPG +T+LL D+ V GLQ R G+ WITV+PV AF++N+GD L
Sbjct: 195 QPDLTLGLSPHSDPGGMTILLPDENVSGLQVRR--GEDWITVKPVPNAFIINMGDQIQVL 368
Query: 269 SNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK-VREGEKSVMEEPITFAE 324
SN +K+ +H+ +VNS+ R+S+A F NP D + P K + ++ + P+TF E
Sbjct: 369 SNAIYKSIEHRVIVNSDKDRVSLAFFYNPRSDIPIQPAKELVTKDRPALYPPMTFDE 539
>TC217748
Length = 1003
Score = 121 bits (303), Expect(2) = 9e-34
Identities = 60/125 (48%), Positives = 81/125 (64%), Gaps = 2/125 (1%)
Frame = +1
Query: 202 VNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQ-VGGLQATRDNGKTWITVQPVEGAFVVN 260
VN+YPKCPQPDLT GL H+DPG +T+LL D V GLQ R G W+ V+PV AFV+N
Sbjct: 337 VNFYPKCPQPDLTFGLSPHSDPGGMTILLSDDFVSGLQVRR--GDEWVIVKPVPNAFVIN 510
Query: 261 LGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK-VREGEKSVMEEP 319
+GD LSN +K+ +H+ +VNSN R+S+A F NP D + P K + E+ + P
Sbjct: 511 IGDQIQVLSNAIYKSVEHRVIVNSNKDRVSLALFYNPRSDLLIQPAKELVTEERPALYSP 690
Query: 320 ITFAE 324
+T+ E
Sbjct: 691 MTYDE 705
Score = 40.4 bits (93), Expect(2) = 9e-34
Identities = 25/99 (25%), Positives = 48/99 (48%)
Frame = +3
Query: 94 EFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTP 153
EFF P E K + + G+ +Q + DW + P R+ ++W P
Sbjct: 3 EFFNQPLEVKEEYANSPTTYEGYGSRLGVQKGATLDWSDYFFLHYRPPSLRNQAKWLAFP 182
Query: 154 AGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKA 192
++ V EY E+++ L ++L+++S +GL+++ L A
Sbjct: 183 QSFRKVIAEYGEEVVKLGGRILKMMSINLGLKEDFLLNA 299
>TC209361 weakly similar to UP|Q7XJ89 (Q7XJ89) Flavonol synthase (Fragment),
partial (19%)
Length = 784
Score = 138 bits (347), Expect = 4e-33
Identities = 78/232 (33%), Positives = 127/232 (54%), Gaps = 6/232 (2%)
Frame = +2
Query: 17 TLESSFVRDEDERPKVAYNNFSNEIPVISLA--GIDEVDGRRSEICNKIVEACENWGIFQ 74
+L F+ EDERP+++ + IP+I L+ D + S + KI +ACE +G FQ
Sbjct: 80 SLSPKFILPEDERPQLSEVTSLDSIPIIDLSDHSYDGNNHSSSLVVQKISQACEEYGFFQ 259
Query: 75 VVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQ---GESVQDWR 131
+V+HG+ ++ + M T + F LPPE+ + T K + + +L GE V+ W
Sbjct: 260 IVNHGIPEQVCNKMMTAITDIFNLPPEQTGQLYTTDHTKNTKLYNYYLNVEGGEKVKMWS 439
Query: 132 EIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTK 191
E +++ YPI + + + + EY+ ++ L +LL +LS +G+E++ L K
Sbjct: 440 ECFSHYWYPIEDIIHLLPQEIGTQYGEAFSEYAREIGSLVRRLLGLLSIGLGIEEDFLLK 619
Query: 192 ACVDMDQ-KVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRD 242
C D + + N+YP CP P+LTLGL HTD +T++LQ QV GLQ +D
Sbjct: 620 ICGDQPRLRAQANFYPPCPDPELTLGLPVHTDFNALTIVLQSQVSGLQVIKD 775
>TC229614 similar to UP|O48631 (O48631) Ethylene-forming-enzyme-like
dioxygenase, partial (52%)
Length = 1289
Score = 128 bits (321), Expect(2) = 6e-33
Identities = 79/245 (32%), Positives = 129/245 (52%), Gaps = 9/245 (3%)
Frame = +1
Query: 91 LAKEFFALPPEEKLRFDMTGGKK-----GGFIVSSHLQGESVQDWREIVTYFSYPIRNRD 145
++K+FF LP EEK + G ++ S Q DW + V P R
Sbjct: 301 VSKQFFQLPKEEKQKCAREREPNNIEGYGNDVIYSKNQR---LDWTDRVYLKVLPEDERK 471
Query: 146 YSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVV-VNY 204
++ WP TP +++ +Y+E L L+ +L+ +++++ LE++ C + +V VNY
Sbjct: 472 FNFWPQTPNDFRSTVLQYTESLRLLSEVILKAMAKSLNLEEDCFLNECGERSNMIVRVNY 651
Query: 205 YPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGD 263
YP CP PD LG+K H D TIT LLQD +V GLQ +D+ W V + A ++N+GD
Sbjct: 652 YPPCPMPDHVLGVKPHADGSTITFLLQDKEVEGLQVLKDD--QWFKVPIIPDALLINVGD 825
Query: 264 HGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPL-KVREGEKSVMEEPI-T 321
+SNG F++ H+ V+N RL++A F P + + P+ K+ + V+ P+
Sbjct: 826 QIEIMSNGIFRSPVHRVVINKAKERLTVAMFCVPDSEKEIKPVDKLVNESRPVLYRPVKN 1005
Query: 322 FAEMY 326
+ E+Y
Sbjct: 1006YVEIY 1020
Score = 30.8 bits (68), Expect(2) = 6e-33
Identities = 14/47 (29%), Positives = 26/47 (54%)
Frame = +3
Query: 39 NEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELV 85
++IPVI L + + E+ K+ A +WG FQ ++HG+ + +
Sbjct: 147 DDIPVIDLHRLSSSSISQQELA-KLHHALHSWGCFQAINHGMKSSFL 284
>TC217747 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (53%)
Length = 713
Score = 137 bits (345), Expect = 8e-33
Identities = 74/182 (40%), Positives = 110/182 (59%), Gaps = 5/182 (2%)
Frame = +1
Query: 148 RWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKAC---VDMDQKVVVNY 204
+W P + V EY E ++ ++L+++S +GL+++ L A ++ + VN+
Sbjct: 1 KWXAFPEXXRKVIAEYGEGVVKXGGRILKMMSINLGLKEDFLLNAFGGESEVGACLRVNF 180
Query: 205 YPKCPQPDLTLGLKRHTDPGTITLLLQDQ-VGGLQATRDNGKTWITVQPVEGAFVVNLGD 263
YPKCPQPDLT GL H+DPG +T+LL D V GLQ R G WITV+PV AF++N+GD
Sbjct: 181 YPKCPQPDLTFGLSPHSDPGGMTILLPDDFVSGLQVRR--GDEWITVKPVPNAFIINIGD 354
Query: 264 HGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK-VREGEKSVMEEPITF 322
LSN +K+ +H+ +VNSN R+S+A F NP D + P K + EK + P+T+
Sbjct: 355 QIQVLSNAIYKSVEHRVIVNSNKDRVSLALFYNPRSDLLIQPAKELVTEEKPALYSPMTY 534
Query: 323 AE 324
E
Sbjct: 535 DE 540
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,831,403
Number of Sequences: 63676
Number of extensions: 194270
Number of successful extensions: 1153
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 1014
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1023
length of query: 366
length of database: 12,639,632
effective HSP length: 99
effective length of query: 267
effective length of database: 6,335,708
effective search space: 1691634036
effective search space used: 1691634036
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0197.2


