

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.5
(903 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
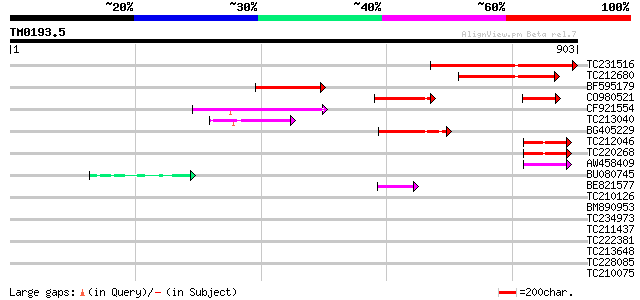
Score E
Sequences producing significant alignments: (bits) Value
TC231516 similar to UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, parti... 323 2e-88
TC212680 similar to UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, parti... 222 5e-58
BF595179 weakly similar to GP|20503001|gb Putataive nodule incep... 175 9e-44
CO980521 155 1e-37
CF921554 152 8e-37
TC213040 similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial (17%) 110 4e-24
BG405229 110 4e-24
TC212046 similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial (9%) 77 4e-14
TC220268 weakly similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial ... 76 8e-14
AW458409 55 1e-07
BU080745 homologue to GP|6448579|emb| nodule inception protein {... 50 6e-06
BE821577 47 5e-05
TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C2... 37 0.039
BM890953 34 0.25
TC234973 34 0.25
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 33 0.73
TC222381 similar to UP|Q76KV2 (Q76KV2) DNA binding with one fing... 32 0.96
TC213648 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide ... 32 1.3
TC228085 similar to UP|Q9LK27 (Q9LK27) Gb|AAF01563.1, partial (17%) 30 4.8
TC210075 homologue to PIR|H96667|H96667 AP2-containing DNA-bindi... 30 4.8
>TC231516 similar to UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (23%)
Length = 835
Score = 323 bits (829), Expect = 2e-88
Identities = 176/236 (74%), Positives = 190/236 (79%), Gaps = 3/236 (1%)
Frame = +2
Query: 671 SNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHS 730
SNFPDLASPNLSG S LNQS+ PNS S QPD G LSPEGA+KS SSSCSQ S+SSHS
Sbjct: 2 SNFPDLASPNLSGTGFFSTLNQSDYPNSTSTQPDHGSLSPEGASKSPSSSCSQSSISSHS 181
Query: 731 CSSMPEQQPH-TSDVACNK-DPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQE 788
CSSM E Q H T++ A NK V +DSA VVLKRI SEAELKS S+++AKL PRS SQE
Sbjct: 182 CSSMSELQQHRTANGAGNKVSTTVSEDSAGVVLKRISSEAELKSLSQDRAKLLPRSQSQE 361
Query: 789 TLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRF 848
TLGEH KT+YQ LLKT ++ K D+HRVKV YGDEKTRFRMPKSW YE LLQEIARRF
Sbjct: 362 TLGEHPKTQYQQPLLKT---SSSKVDSHRVKVAYGDEKTRFRMPKSWGYEDLLQEIARRF 532
Query: 849 NVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS-GMRS 903
NVSDMSKFDVKYLDDD EWVLLTCDADLEECIDVC SSES TIKL +Q SS MRS
Sbjct: 533 NVSDMSKFDVKYLDDDCEWVLLTCDADLEECIDVCQSSESGTIKLSLQPSSHSMRS 700
>TC212680 similar to UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (13%)
Length = 479
Score = 222 bits (566), Expect = 5e-58
Identities = 118/162 (72%), Positives = 136/162 (83%), Gaps = 2/162 (1%)
Frame = +1
Query: 716 SLSSSCSQGSLSSHSCSSMPE-QQPHTSDVACNKDPV-VGKDSADVVLKRIRSEAELKSH 773
S SSS SQ S+SSHSCSSM E QQ HT+++A +KDP VG+ SADVVLK IR+EA+LKS
Sbjct: 1 SPSSSSSQSSISSHSCSSMSELQQQHTTNIASDKDPATVGEYSADVVLKLIRNEAKLKSL 180
Query: 774 SENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPK 833
S+++AKL PRSLSQETLGEH KT+YQ LLKT ++ K D+HRVKVTYGDEKTRFRM K
Sbjct: 181 SQDRAKLLPRSLSQETLGEHPKTQYQLPLLKT---SSSKVDSHRVKVTYGDEKTRFRMLK 351
Query: 834 SWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDAD 875
+W YE LLQEI R+FNVSDMSKFDVKYLDDD EW+LLTCDAD
Sbjct: 352 NWVYEDLLQEIGRKFNVSDMSKFDVKYLDDDCEWILLTCDAD 477
>BF595179 weakly similar to GP|20503001|gb Putataive nodule inception protein
{Oryza sativa (japonica cultivar-group)}, partial (11%)
Length = 360
Score = 175 bits (443), Expect = 9e-44
Identities = 88/112 (78%), Positives = 96/112 (85%)
Frame = +2
Query: 392 EHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSA 451
EHHLLKG+GVAG AFM NQP FS DIT LSK DYP+SHHARLFGLRAAVAIRLRSIY+S
Sbjct: 14 EHHLLKGEGVAGGAFMTNQPCFSDDITSLSKKDYPMSHHARLFGLRAAVAIRLRSIYNST 193
Query: 452 DDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSSSV 503
DD+VLEFFLPV+CND EEQ+ ML SLS IIQR CRSLRVI +KELE + SV
Sbjct: 194 DDFVLEFFLPVDCNDIEEQRKMLTSLSNIIQRVCRSLRVIREKELEEANLSV 349
>CO980521
Length = 657
Score = 155 bits (391), Expect = 1e-37
Identities = 77/97 (79%), Positives = 87/97 (89%)
Frame = -2
Query: 582 KTGEKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKK 641
K GEKRR KA+KTI+L VLRQYF GSLKDAAK+IGVC TTLKR+CRQHGI RWPSRKIKK
Sbjct: 563 KPGEKRRTKAEKTISLPVLRQYFAGSLKDAAKSIGVCPTTLKRICRQHGITRWPSRKIKK 384
Query: 642 VGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLAS 678
VGHSL+KLQLVIDSVQGA GA +I SFY++FP+L+S
Sbjct: 383 VGHSLKKLQLVIDSVQGAEGA-IQIGSFYNSFPELSS 276
Score = 72.8 bits (177), Expect = 6e-13
Identities = 35/63 (55%), Positives = 43/63 (67%), Gaps = 2/63 (3%)
Frame = -2
Query: 817 RVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSK--FDVKYLDDDLEWVLLTCDA 874
RVK T+ DEK RF + W + L EIARRFN++D+S +KYLDDD EWV+L CD
Sbjct: 191 RVKATFADEKIRFSLQPHWGFTELQLEIARRFNLNDVSNGYLVLKYLDDDGEWVVLACDG 12
Query: 875 DLE 877
DLE
Sbjct: 11 DLE 3
>CF921554
Length = 812
Score = 152 bits (383), Expect = 8e-37
Identities = 89/226 (39%), Positives = 131/226 (57%), Gaps = 10/226 (4%)
Frame = -1
Query: 291 LESVCKALEVVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC 350
++ +CKALE V+ S + + ++ + AL EI E+L C LPLAQTW+ C
Sbjct: 812 VDKICKALETVNXRSSEILDHPYTQICNEGRQNALSEILEILTVVCETLNLPLAQTWIPC 633
Query: 351 -----FQQG---KDGCRHSEDNYLH--CISPVEQACYVGDPSVRFFHEACMEHHLLKGQG 400
QG K C + + + C+S + A Y+ D + F EAC+EHHL +GQG
Sbjct: 632 KHRSVLAQGGGVKKSCSSFDGSCMGKVCMSTTDIAFYIIDAHLWGFREACVEHHLQQGQG 453
Query: 401 VAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFL 460
VAG+AF+ + F ++IT KTDYPL H+A +FGL + I LRS ++ DDYVLEFFL
Sbjct: 452 VAGRAFLSHSMCFCSNITQFCKTDYPLVHYALMFGLTSCFTICLRSSHTGNDDYVLEFFL 273
Query: 461 PVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSSSVEVM 506
P D EQK +L S+ I+++ +SL++ + ELE S+E++
Sbjct: 272 PPRITDFHEQKTLLGSILAIMKQHFQSLKIASGVELE--DGSIEII 141
>TC213040 similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial (17%)
Length = 466
Score = 110 bits (274), Expect = 4e-24
Identities = 64/152 (42%), Positives = 83/152 (54%), Gaps = 14/152 (9%)
Frame = +3
Query: 318 DKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGK--------------DGCRHSED 363
D+ Y AL EI +L C H LPLAQTWV C + DGC +
Sbjct: 3 DRQY--ALAEI*SILTVVCETHSLPLAQTWVPCKHRSVLAHGGGHKKSCSSFDGCCMGQV 176
Query: 364 NYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKT 423
C+S E A YV D FHEAC+EHHL +GQGVAG+AF+ + F +I KT
Sbjct: 177 ----CMSITEVAFYVIDAHTWGFHEACVEHHLQQGQGVAGRAFLSHNMCFCGNIAQFCKT 344
Query: 424 DYPLSHHARLFGLRAAVAIRLRSIYSSADDYV 455
+YPL H+A +FGL + A+ L+S ++ DDYV
Sbjct: 345 EYPLVHYALMFGLTSCFAVCLQSSHTGNDDYV 440
>BG405229
Length = 382
Score = 110 bits (274), Expect = 4e-24
Identities = 59/116 (50%), Positives = 80/116 (68%)
Frame = +1
Query: 588 RAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQ 647
R +A+ +I+L VL+ YF GSLKDAAK++GVC TT+KR+CRQHGI RWPS KIKKV HSL
Sbjct: 7 RHEAEISISLNVLQHYFTGSLKDAAKSLGVCPTTMKRICRQHGISRWPSLKIKKVNHSLS 186
Query: 648 KLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQP 703
KL+ VI+SV GA GA F ++S + +A+ + S N+ +L+I+P
Sbjct: 187 KLKCVIESVHGAEGA-FGLNSLSTGSLPIAAGSFSDP---FTSNKFHRQTTLTIRP 342
>TC212046 similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial (9%)
Length = 977
Score = 76.6 bits (187), Expect = 4e-14
Identities = 36/78 (46%), Positives = 54/78 (69%)
Frame = +3
Query: 818 VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLE 877
+K TY ++ RFR+ + L +EIA+R + ++ FD+KYLDDD EWVL+ CDADL+
Sbjct: 531 IKATYREDIIRFRVSLTCGIVELKEEIAKRLKL-EVGTFDIKYLDDDHEWVLIACDADLQ 707
Query: 878 ECIDVCLSSESSTIKLCI 895
EC+DV SS S+ I++ +
Sbjct: 708 ECMDVSRSSGSNIIRVLV 761
>TC220268 weakly similar to UP|Q9SGW2 (Q9SGW2) F1N19.10, partial (8%)
Length = 857
Score = 75.9 bits (185), Expect = 8e-14
Identities = 35/78 (44%), Positives = 52/78 (65%)
Frame = +3
Query: 818 VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLE 877
+K TY ++ RF++ L +EIA+R + + FD+KYLDDD EWVL+ CDADL+
Sbjct: 507 IKATYKEDIIRFKVSMDCGIVELKEEIAKRLKL-EAGTFDIKYLDDDHEWVLIACDADLQ 683
Query: 878 ECIDVCLSSESSTIKLCI 895
EC+D+ SS S+ I+L +
Sbjct: 684 ECMDISRSSGSNVIRLVV 737
>AW458409
Length = 292
Score = 55.5 bits (132), Expect = 1e-07
Identities = 27/78 (34%), Positives = 43/78 (54%)
Frame = +2
Query: 818 VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLE 877
+ TY + F + L +E+A +N+ FD+ LDDD EWVL+ CDADL
Sbjct: 20 ISATYKEYVIEFSVSMDCGIVALKEEVAVGWNLQ-AGAFDIYCLDDDHEWVLIACDADLL 196
Query: 878 ECIDVCLSSESSTIKLCI 895
+C+D+ SS + ++L +
Sbjct: 197 DCMDISRSSRXNVVRLVV 250
>BU080745 homologue to GP|6448579|emb| nodule inception protein {Lotus
japonicus}, partial (2%)
Length = 429
Score = 49.7 bits (117), Expect = 6e-06
Identities = 42/168 (25%), Positives = 66/168 (39%)
Frame = +2
Query: 128 KRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPF 187
K WWI P + S+ E+L A+ +++++ N IQIWVP+ R L +
Sbjct: 20 KTWWIGPKANHA---SVKERLEAAMGYLREYTNNN---IQIWVPLRRSAGQELGTDE--- 172
Query: 188 SLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPR 247
S+ F + K +R FRS E
Sbjct: 173 ---------------SDTIAFERNRNVK---------------------LRLFRSQE--- 235
Query: 248 VEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVC 295
G + VP+ E+GS TCLGV+E+VM + ++ + + VC
Sbjct: 236 ----------GCVGVPVLERGSGTCLGVLEIVMEDEVVSEMMXVVKVC 349
>BE821577
Length = 555
Score = 46.6 bits (109), Expect = 5e-05
Identities = 27/66 (40%), Positives = 36/66 (53%), Gaps = 2/66 (3%)
Frame = -3
Query: 587 RRAKADKT--ITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGH 644
+RA +D I L L F + +A++N V T LKR CR+ GI RWP RKIK +
Sbjct: 553 KRAXSDLVAKIXLSDLVXXFGMPIVEASRNXNVGLTVLKRKCREFGIPRWPHRKIKSLDS 374
Query: 645 SLQKLQ 650
+ LQ
Sbjct: 373 LIHDLQ 356
>TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23), partial
(7%)
Length = 735
Score = 37.0 bits (84), Expect = 0.039
Identities = 29/97 (29%), Positives = 45/97 (45%), Gaps = 2/97 (2%)
Frame = -3
Query: 639 IKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNS 698
++ + + ++ + +S +S S S+ P +S +LS +S S+L S
Sbjct: 634 LETISYEYSHFLFLVGGCEVSSSSSSSSSSSSSSSPSSSSSSLSLSSSSSSLPPPLASGS 455
Query: 699 LSIQPDLGPLSPEGATKSLSS--SCSQGSLSSHSCSS 733
S P P SPE ++ S SS SC S SS S SS
Sbjct: 454 SSGSPSSSPSSPENSSSSPSSS*SCPSSSSSSPSLSS 344
>BM890953
Length = 433
Score = 34.3 bits (77), Expect = 0.25
Identities = 16/66 (24%), Positives = 35/66 (52%)
Frame = +3
Query: 838 EHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQA 897
+ L+ I +R +V+D+ + + Y DD+ + ++L D+DL + S+ +KL +
Sbjct: 6 DELVSTIMQRLDVNDVERPMILYEDDEGDKIVLATDSDLVSAVSYATSAGMKALKLHLDF 185
Query: 898 SSGMRS 903
S ++
Sbjct: 186 GSSFKA 203
>TC234973
Length = 384
Score = 34.3 bits (77), Expect = 0.25
Identities = 33/95 (34%), Positives = 44/95 (45%)
Frame = +1
Query: 639 IKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNS 698
I+ HSL L +D + +SG S + + S L+G S S N S PNS
Sbjct: 4 IRHEDHSLHGAHLNLDIQKISSGNSLNVST---------SRELNGVSSASKDNLS--PNS 150
Query: 699 LSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSS 733
+ LSP GAT+S S S SLS++S S
Sbjct: 151 RVV------LSPVGATRSTSISAPSMSLSNNSTPS 237
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 32.7 bits (73), Expect = 0.73
Identities = 31/85 (36%), Positives = 41/85 (47%), Gaps = 7/85 (8%)
Frame = -1
Query: 656 VQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSL-------SIQPDLGPL 708
V G G S S S+ +S S +S +S+ + S +P+SL S P P
Sbjct: 626 VGGGCGVSSSSSSSSSSSSSPSSS--SSSSPLSSPSSSSSPSSLPSPLASSSGSPPSSPS 453
Query: 709 SPEGATKSLSSSCSQGSLSSHSCSS 733
SPE ++ S SSS S S SS S SS
Sbjct: 452 SPENSSSSPSSSSSGPSSSSSSPSS 378
>TC222381 similar to UP|Q76KV2 (Q76KV2) DNA binding with one finger 2
protein, partial (36%)
Length = 485
Score = 32.3 bits (72), Expect = 0.96
Identities = 16/47 (34%), Positives = 28/47 (59%)
Frame = +2
Query: 691 NQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQ 737
NQ + N L PD+ P +P AT ++++ SQG+ +SH+ P++
Sbjct: 128 NQRLSQNKL*SAPDVTPPTPNSATTTITAFPSQGTSASHAGDIGPKE 268
>TC213648 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide
repeat(TPR)-containing protein-like, partial (19%)
Length = 727
Score = 32.0 bits (71), Expect = 1.3
Identities = 17/47 (36%), Positives = 29/47 (61%)
Frame = +2
Query: 653 IDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSL 699
+DSV+ S K+ + + ++PD+AS + S++SALN S + SL
Sbjct: 587 VDSVKTWLYPSLKVTTSWKDYPDIASTFATTGSVISALNFSSDD*SL 727
>TC228085 similar to UP|Q9LK27 (Q9LK27) Gb|AAF01563.1, partial (17%)
Length = 947
Score = 30.0 bits (66), Expect = 4.8
Identities = 18/42 (42%), Positives = 23/42 (53%)
Frame = -1
Query: 691 NQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCS 732
NQS +P+S S GP +P G S SSS S + + SCS
Sbjct: 485 NQSISPSSTSFD---GPAAPLGGGSSSSSSSSSSFMYNPSCS 369
>TC210075 homologue to PIR|H96667|H96667 AP2-containing DNA-binding protein,
51686-52693 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (40%)
Length = 1024
Score = 30.0 bits (66), Expect = 4.8
Identities = 34/137 (24%), Positives = 55/137 (39%), Gaps = 27/137 (19%)
Frame = +2
Query: 618 CTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGAS----GASFKIDSFYS-- 671
C L R RQ +W + +I+ + ++ D+ + A+ A++K+ Y+
Sbjct: 221 CKKKLYRGVRQRHWGKWVA-EIRLPQNRMRVWLGTYDTAEAAAYAYDRAAYKLRGEYARL 397
Query: 672 NFPDLASPNLSG---ASLVSALNQS------------------ENPNSLSIQPDLGPLSP 710
NFP+L P G ++ ++AL S +N + G P
Sbjct: 398 NFPNLKDPTKLGFGDSARLNALKSSVDAKIQAICQKVKKERAKKNATKKKLHDSSGSTDP 577
Query: 711 EGATKSLSSSCSQGSLS 727
G K SSSCS SLS
Sbjct: 578 NGGQKINSSSCSSSSLS 628
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,992,708
Number of Sequences: 63676
Number of extensions: 631757
Number of successful extensions: 3449
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 3401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3437
length of query: 903
length of database: 12,639,632
effective HSP length: 106
effective length of query: 797
effective length of database: 5,889,976
effective search space: 4694310872
effective search space used: 4694310872
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0193.5


