

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
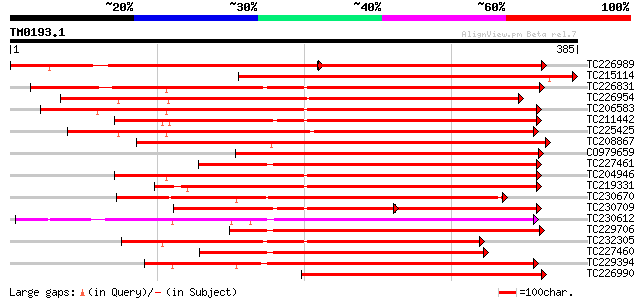
Query= TM0193.1
(385 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 233 e-118
TC215114 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), parti... 385 e-107
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 334 3e-92
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 312 1e-85
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 308 2e-84
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 303 7e-83
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 300 6e-82
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 293 9e-80
CO979659 287 5e-78
TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein ... 284 4e-77
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 280 8e-76
TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like prote... 277 5e-75
TC230670 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 271 5e-73
TC230709 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 187 1e-70
TC230612 similar to GB|BAA98102.1|8978074|AB025609 protein serin... 262 2e-70
TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 261 4e-70
TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase... 260 8e-70
TC227460 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 257 5e-69
TC229394 similar to GB|BAA98102.1|8978074|AB025609 protein serin... 250 9e-67
TC226990 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 250 9e-67
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 233 bits (593), Expect(2) = e-118
Identities = 109/155 (70%), Positives = 133/155 (85%)
Frame = +3
Query: 210 RDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKS 269
RD KS+NILLD ++PKLSDFGLAKLGPVGD +HVSTRVMGTYGYCAPEYAM+G+LT+KS
Sbjct: 660 RDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKS 839
Query: 270 DIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRC 329
D+YSFGVV LEL+TGR+AID+++ GEQNLV+WARP F+DRR+F + DP LQGRFP R
Sbjct: 840 DVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRLQGRFPMRG 1019
Query: 330 LHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
L+QA+A+ +MC+QE RPLI D+V AL YLA+Q
Sbjct: 1020LYQALAVASMCIQESAATRPLIGDVVTALSYLANQ 1124
Score = 211 bits (536), Expect(2) = e-118
Identities = 116/217 (53%), Positives = 146/217 (66%), Gaps = 4/217 (1%)
Frame = +2
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATR---SGSSSGNGGVNAAAAATDGERKRETKGKGKGV 57
M CFSC S K+ + + S S G + + ++G KRE + V
Sbjct: 47 MGCFSCFDSSSKEDHNLRPQHQPNQPLPSQISRLPSGADKLRSRSNGGSKRELQQPPPTV 226
Query: 58 SSSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLS 116
+ AA +F FRELA AT+NF+ + +GEGGFG+VYKGRL TT + VAVKQL
Sbjct: 227 ---------QIAAQTFTFRELAAATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLD 379
Query: 117 HDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHD 176
+G QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYE+MP+GSLEDHL +L D
Sbjct: 380 KNGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPD 559
Query: 177 KEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLK 213
KEPL+W+TRMK+AVGAA+GLEYLH A+PP I + L+
Sbjct: 560 KEPLDWNTRMKIAVGAAKGLEYLHDKANPPGIQQRLQ 670
>TC215114 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (54%)
Length = 878
Score = 385 bits (990), Expect = e-107
Identities = 191/233 (81%), Positives = 207/233 (87%), Gaps = 3/233 (1%)
Frame = +1
Query: 156 LVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSA 215
LVYEYMPMGSLEDHLF+ DKEPL+WSTRMK+AVGAARGLEYLHC ADPPVIYRDLKSA
Sbjct: 1 LVYEYMPMGSLEDHLFDPHPDKEPLSWSTRMKIAVGAARGLEYLHCKADPPVIYRDLKSA 180
Query: 216 NILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFG 275
NILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFG
Sbjct: 181 NILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFG 360
Query: 276 VVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIA 335
V+LLEL+TGRRAIDT+RRPGEQNLVSW+R +FSDR++F M+DPLLQ FP RCL+QA+A
Sbjct: 361 VLLLELITGRRAIDTNRRPGEQNLVSWSRQFFSDRKKFVQMIDPLLQENFPLRCLNQAMA 540
Query: 336 ITAMCLQEQPKFRPLITDIVVALEYLASQG---SPEAHYLYGVQQRPSRMERN 385
ITAMC+QEQPKFRPLI DIVVALEYLAS SP + QQ PS M+ N
Sbjct: 541 ITAMCIQEQPKFRPLIGDIVVALEYLASHSNP*SP*TWWCSSPQQPPSEMDIN 699
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 334 bits (857), Expect = 3e-92
Identities = 181/359 (50%), Positives = 234/359 (64%), Gaps = 10/359 (2%)
Frame = +1
Query: 15 SRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGSSNGKTAAASFG 74
SR + T S N + + + A+D + +G+ +SS N +F
Sbjct: 289 SRTPSGISKTSPSSVPSNLSILSYSEASDFSNLPTPRSEGEILSSPN--------LKAFT 444
Query: 75 FRELADATRNFKEANLIGEGGFGKVYKGRLT----------TGEAVAVKQLSHDGRQGFQ 124
F EL +ATRNF+ +L+GEGGFG VYKG + +G VAVK+L +G QG +
Sbjct: 445 FNELKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGLQGHK 624
Query: 125 EFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWST 184
E++ EV L LHH NLV+LIGYC DG+ RLLVYE+M GSLE+HLF +PL+WS
Sbjct: 625 EWLTEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFR--RGPQPLSWSV 798
Query: 185 RMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHV 244
RMKVA+GAARGL +LH A VIYRD K++NILLD EFN KLSDFGLAK GP GD THV
Sbjct: 799 RMKVAIGAARGLSFLH-NAKSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGDRTHV 975
Query: 245 STRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWAR 304
ST+VMGT GY APEY +G+LT KSD+YSFGVVLLELL+GRRA+D S+ EQNLV WA+
Sbjct: 976 STQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLVEWAK 1155
Query: 305 PYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLAS 363
PY D+RR ++D L G++P + + A + CL + K RP IT+++ LE +A+
Sbjct: 1156PYLGDKRRLFRIMDTKLGGQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTLEQIAA 1332
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 312 bits (800), Expect = 1e-85
Identities = 161/339 (47%), Positives = 217/339 (63%), Gaps = 24/339 (7%)
Frame = +2
Query: 35 VNAAAAATDGERKRETKGKGKGVSSSNGSSNGKTAAAS--------------FGFRELAD 80
+ A + + G R G +SS++ SS+ S F + EL
Sbjct: 149 IKAVSPSNTGITSRSVSRSGHDISSNSRSSSASIPVTSRSEGEILQSSNLKSFSYHELRA 328
Query: 81 ATRNFKEANLIGEGGFGKVYKGRLTT----------GEAVAVKQLSHDGRQGFQEFVMEV 130
ATRNF+ +++GEGGFG V+KG + G+ VAVK+L+ DG QG +E++ E+
Sbjct: 329 ATRNFRPDSVLGEGGFGSVFKGWIDEHSLAATKPGIGKIVAVKKLNQDGLQGHREWLAEI 508
Query: 131 LMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAV 190
L L H NLV+LIGYC + + RLLVYE+MP GS+E+HLF +P +WS RMK+A+
Sbjct: 509 NYLGQLQHPNLVKLIGYCFEDEHRLLVYEFMPKGSMENHLFRRGSYFQPFSWSLRMKIAL 688
Query: 191 GAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMG 250
GAA+GL +LH T + VIYRD K++NILLD +N KLSDFGLA+ GP GD +HVSTRVMG
Sbjct: 689 GAAKGLAFLHST-EHKVIYRDFKTSNILLDTNYNAKLSDFGLARDGPTGDKSHVSTRVMG 865
Query: 251 TYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDR 310
T GY APEY +G LT KSD+YSFGVVLLE+++GRRAID ++ GE NLV WA+PY S++
Sbjct: 866 TRGYAAPEYLATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHNLVEWAKPYLSNK 1045
Query: 311 RRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRP 349
RR ++DP L+G++ A A+ C +PK RP
Sbjct: 1046RRVFRVMDPRLEGQYSQNRAQAAAALAMQCFSVEPKCRP 1162
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 308 bits (790), Expect = 2e-84
Identities = 164/355 (46%), Positives = 227/355 (63%), Gaps = 15/355 (4%)
Frame = +1
Query: 22 NATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVS-----SSNGSSNGKTAAASFGFR 76
+A S N G N+ +TDG T K S S G + SF
Sbjct: 76 SAQIKAESPYNTGFNSKYVSTDGNDLGSTNDKVSANSIPQTPRSEGEILQSSNLKSFTLS 255
Query: 77 ELADATRNFKEANLIGEGGFGKVYKGRLT----------TGEAVAVKQLSHDGRQGFQEF 126
EL ATRNF+ +++GEGGFG V+KG + TG +AVK+L+ DG G +E+
Sbjct: 256 ELKTATRNFRPDSVLGEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQDGIPGHREW 435
Query: 127 VMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRM 186
+ EV L H++LVRLIG+C + + RLLVYE+MP GS E+HLF +PL+WS R+
Sbjct: 436 LAEVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFRRGSYFQPLSWSLRL 615
Query: 187 KVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST 246
KVA+ AA+GL +LH +A+ VIYRD K++N+LLD+++N KLSDFGLAK GP GD +HVST
Sbjct: 616 KVALDAAKGLAFLH-SAEAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGDKSHVST 792
Query: 247 RVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPY 306
RVMGTYGY APEY +G LT KSD+YSFGVVLLE+L+G+RA+D +R G+ NLV WA+P+
Sbjct: 793 RVMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLVEWAKPF 972
Query: 307 FSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
+++R+ ++D LQG++ + ++ + CL + KFRP + +V LE L
Sbjct: 973 MANKRKIFRVLDTRLQGQYSTDDAYKLATLALRCLSIESKFRPNMDQVVTTLEQL 1137
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 303 bits (777), Expect = 7e-83
Identities = 155/297 (52%), Positives = 209/297 (70%), Gaps = 7/297 (2%)
Frame = +2
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKG----RLTTG---EAVAVKQLSHDGRQGFQ 124
+F EL +AT +F +N++GEGGFG VYKG +L +G + VAVK+L DG QG +
Sbjct: 32 AFTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLDLDGLQGHR 211
Query: 125 EFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWST 184
E++ E++ L L H +LV+LIGYC + + RLL+YEYMP GSLE+ LF P WST
Sbjct: 212 EWLAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYSAAMP--WST 385
Query: 185 RMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHV 244
RMK+A+GAA+GL +LH AD PVIYRD K++NILLD++F KLSDFGLAK GP G++THV
Sbjct: 386 RMKIALGAAKGLAFLH-EADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDGPEGEDTHV 562
Query: 245 STRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWAR 304
+TR+MGT GY APEY M+G LT KSD+YS+GVVLLELLTGRR +D SR ++LV WAR
Sbjct: 563 TTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGKSLVEWAR 742
Query: 305 PYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
P D+++ +++D L+G+FP + + + CL P RP ++D++ LE L
Sbjct: 743 PLLRDQKKVYNIIDRRLEGQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVIKVLEPL 913
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 300 bits (769), Expect = 6e-82
Identities = 157/332 (47%), Positives = 210/332 (62%), Gaps = 12/332 (3%)
Frame = +1
Query: 40 AATDGERKRETKGKGKGVSSSNGSSNGKTAAAS----FGFRELADATRNFKEANLIGEGG 95
A T R R K S NG+ S F F EL ATRNFK ++GEGG
Sbjct: 1 ADTYRNRNRYINSKFSVSSGDQPYPNGQILPTSNLRIFTFAELKAATRNFKADTVLGEGG 180
Query: 96 FGKVYKGRLT--------TGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGY 147
FGKV+KG L +G +AVK+L+ + QG +E+ EV L L HTNLV+L+GY
Sbjct: 181 FGKVFKGWLEEKATSKGGSGTVIAVKKLNSESLQGLEEWQSEVNFLGRLSHTNLVKLLGY 360
Query: 148 CTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPV 207
C + + LLVYE+M GSLE+HLF +PL W R+K+A+GAARGL +LH + V
Sbjct: 361 CLEESELLLVYEFMQKGSLENHLFGRGSAVQPLPWDIRLKIAIGAARGLAFLHTSEK--V 534
Query: 208 IYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTL 267
IYRD K++NILLD +N K+SDFGLAKLGP +HV+TRVMGT+GY APEY +G L +
Sbjct: 535 IYRDFKASNILLDGSYNAKISDFGLAKLGPSASQSHVTTRVMGTHGYAAPEYVATGHLYV 714
Query: 268 KSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPS 327
KSD+Y FGVVL+E+LTG RA+D++R G+ L W +PY DRR+ ++D L+G+FPS
Sbjct: 715 KSDVYGFGVVLVEILTGLRALDSNRPSGQHKLTEWVKPYLHDRRKLKGIMDSRLEGKFPS 894
Query: 328 RCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
+ + ++ CL +PK RP + D++ LE
Sbjct: 895 KAAFRIAQLSMKCLASEPKHRPSMKDVLENLE 990
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 293 bits (750), Expect = 9e-80
Identities = 149/285 (52%), Positives = 203/285 (70%), Gaps = 4/285 (1%)
Frame = +1
Query: 87 EANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIG 146
++N+IG GGFG VY+G L G VA+K + G+QG +EF +EV +LS LH L+ L+G
Sbjct: 4 KSNVIGHGGFGLVYRGVLNDGRKVAIKFMDQAGKQGEEEFKVEVELLSRLHSPYLLALLG 183
Query: 147 YCTDGDQRLLVYEYMPMGSLEDHLFELSHD---KEPLNWSTRMKVAVGAARGLEYLHCTA 203
YC+D + +LLVYE+M G L++HL+ +S+ L+W TR+++A+ AA+GLEYLH
Sbjct: 184 YCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLHEHV 363
Query: 204 DPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSG 263
PPVI+RD KS+NILLD +F+ K+SDFGLAKLGP HVSTRV+GT GY APEYA++G
Sbjct: 364 SPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALTG 543
Query: 264 KLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQG 323
LT KSD+YS+GVVLLELLTGR +D R PGE LVSWA P +DR + ++DP L+G
Sbjct: 544 HLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPSLEG 723
Query: 324 RFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA-SQGSP 367
++ + + Q AI AMC+Q + +RPL+ D+V +L L +Q SP
Sbjct: 724 QYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSLVPLVKTQRSP 858
>CO979659
Length = 731
Score = 287 bits (735), Expect = 5e-78
Identities = 136/209 (65%), Positives = 166/209 (79%)
Frame = -1
Query: 154 RLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLK 213
R+LVYE+M GSLE+HL ++ DKEP++W RMK+A GAARGLEYLH ADP +IYRD K
Sbjct: 725 RILVYEFMSNGSLENHLLDIGADKEPMDWKNRMKIAEGAARGLEYLHNGADPAIIYRDFK 546
Query: 214 SANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYS 273
S+NILLD FNPKLSDFGLAK+GP HV+TRVMGT+GYCAPEYA SG+L+ KSDIYS
Sbjct: 545 SSNILLDENFNPKLSDFGLAKIGPKEGEEHVATRVMGTFGYCAPEYAASGQLSTKSDIYS 366
Query: 274 FGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQA 333
FGVVLLE++TGRR DT+R EQNL+ WA+P F DR +F M DPLL+G+FP + L QA
Sbjct: 365 FGVVLLEIITGRRVFDTARGTEEQNLIDWAQPLFKDRTKFTLMADPLLKGQFPVKGLFQA 186
Query: 334 IAITAMCLQEQPKFRPLITDIVVALEYLA 362
+A+ AMCLQE+P RP + D+V AL +LA
Sbjct: 185 LAVAAMCLQEEPDTRPYMDDVVTALAHLA 99
>TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (59%)
Length = 1204
Score = 284 bits (727), Expect = 4e-77
Identities = 143/233 (61%), Positives = 176/233 (75%)
Frame = +2
Query: 129 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 188
EV L L H +LV+LIGYC + DQRLLVYE+MP GSLE+HLF S PL WS RMK+
Sbjct: 8 EVNYLGDLVHPHLVKLIGYCIEDDQRLLVYEFMPRGSLENHLFRRSL---PLPWSIRMKI 178
Query: 189 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 248
A+GAA+GL +LH A+ PVIYRD K++NILLD E+N KLSDFGLAK GP GD THVSTRV
Sbjct: 179 ALGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFGLAKDGPEGDKTHVSTRV 358
Query: 249 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFS 308
MGTYGY APEY M+G LT +SD+YSFGVVLLE+LTGRR++D +R GE NLV WARP+
Sbjct: 359 MGTYGYAAPEYVMTGHLTSRSDVYSFGVVLLEMLTGRRSMDKNRPNGEHNLVEWARPHLG 538
Query: 309 DRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
+RRRF ++DP L+G F + +A + A CL PK RPL++++V AL+ L
Sbjct: 539 ERRRFYKLIDPRLEGHFSIKGAQKAAHLAAHCLSRDPKARPLMSEVVEALKPL 697
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 280 bits (716), Expect = 8e-76
Identities = 145/301 (48%), Positives = 203/301 (67%), Gaps = 11/301 (3%)
Frame = +3
Query: 72 SFGFRELADATRNFKEANLIG-EGGFGKVYKGRLT----------TGEAVAVKQLSHDGR 120
+F EL ATRNF++ +++G EG FG V+KG + TG VAVK+LS D
Sbjct: 303 NFSLTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKRLSLDSF 482
Query: 121 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 180
QG ++ + EV L L H +LV+LIGYC + RLLVYE+MP GSLE+HLF +PL
Sbjct: 483 QGHKDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLFMRGSYFQPL 662
Query: 181 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 240
+W R+KVA+GAA+GL +LH +A+ VIYRD K++N+LLD+ +N KL+D GLAK GP +
Sbjct: 663 SWGLRLKVALGAAKGLAFLH-SAETKVIYRDFKTSNVLLDSNYNAKLADLGLAKDGPTRE 839
Query: 241 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 300
+H STRVMGTYGY APEY +G L+ KSD++SFGVVLLE+L+GRRA+D +R G+ NLV
Sbjct: 840 KSHASTRVMGTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVDKNRPSGQHNLV 1019
Query: 301 SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 360
WA+PY S++R+ ++D L+G++ + ++ CL + K RP + ++ LE
Sbjct: 1020EWAKPYLSNKRKLLRVLDNRLEGQYELDEACKVATLSLRCLAIESKLRPTMDEVATDLEQ 1199
Query: 361 L 361
L
Sbjct: 1200L 1202
>TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like protein
(At5g56460), partial (61%)
Length = 1183
Score = 277 bits (709), Expect = 5e-75
Identities = 142/266 (53%), Positives = 187/266 (69%), Gaps = 3/266 (1%)
Frame = +1
Query: 99 VYKGRLTTGEAVAVKQLSHDG---RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRL 155
+ KG T AV V HDG QG +E++ +V L H NLV++IGYC + + R+
Sbjct: 37 IRKGLPTLDVAVKV----HDGDNSHQGHREWLSQVEFWGQLSHPNLVKVIGYCCEDNHRV 204
Query: 156 LVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSA 215
L+YEYM G L+++LF+ + PL+WS RMK+A GAA+GL +LH A+ PVIYRD K++
Sbjct: 205 LIYEYMSRGGLDNYLFKYAPAIPPLSWSMRMKIAFGAAKGLAFLH-EAEKPVIYRDFKTS 381
Query: 216 NILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFG 275
NILLD E+N KLSDFGLAK GPVGD +HVSTRVMGTYGY APEY M+G LT +SD+YSFG
Sbjct: 382 NILLDQEYNSKLSDFGLAKDGPVGDKSHVSTRVMGTYGYAAPEYIMTGHLTPRSDVYSFG 561
Query: 276 VVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIA 335
VVLLELLTGR+++D R EQNL WA P ++++F +++DP L G +P + +H+A
Sbjct: 562 VVLLELLTGRKSLDKLRPAREQNLAEWALPLLKEKKKFLNIIDPRLDGDYPIKAVHKAAM 741
Query: 336 ITAMCLQEQPKFRPLITDIVVALEYL 361
+ CL PK RPL+ DIV +LE L
Sbjct: 742 LAYHCLNRNPKARPLMRDIVDSLEPL 819
>TC230670 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (10%)
Length = 1094
Score = 271 bits (692), Expect = 5e-73
Identities = 141/270 (52%), Positives = 187/270 (69%), Gaps = 4/270 (1%)
Frame = +2
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 132
F F +L ATR F A L+GEGGFG VY+G L + VA+KQL+ +G QG +E++ E+ +
Sbjct: 287 FSFSDLKSATRAFSRALLVGEGGFGSVYRGLLDQND-VAIKQLNRNGHQGHKEWINELNL 463
Query: 133 LSLLHHTNLVRLIGYCTDGD----QRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 188
L ++ H NLV+L+GYC + D QRLLVYE+MP SLEDHL + W TR+++
Sbjct: 464 LGVVKHPNLVKLVGYCAEDDERGIQRLLVYEFMPNKSLEDHLLARV-PSTIIPWGTRLRI 640
Query: 189 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 248
A AARGL YLH D +I+RD K++NILLD FN KLSDFGLA+ GP + +VST V
Sbjct: 641 AQDAARGLAYLHEEMDFQLIFRDFKTSNILLDENFNAKLSDFGLARQGPSEGSGYVSTAV 820
Query: 249 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFS 308
+GT GY APEY +GKLT KSD++SFGVVL EL+TGRRA+D + EQ L+ W RPY S
Sbjct: 821 VGTIGYAAPEYVQTGKLTAKSDVWSFGVVLYELITGRRAVDRNLPRNEQKLLDWVRPYVS 1000
Query: 309 DRRRFGHMVDPLLQGRFPSRCLHQAIAITA 338
D R+F H++DP L+G++ + H+ +AI A
Sbjct: 1001DPRKFHHILDPRLKGQYCIKSAHK-LAILA 1087
>TC230709 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (61%)
Length = 1245
Score = 187 bits (474), Expect(2) = 1e-70
Identities = 95/153 (62%), Positives = 116/153 (75%)
Frame = +3
Query: 112 VKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLF 171
VK L DG QG +E++ EV+ L L H +LV+LIGYC + + RLLVYEY+P GSLE+ LF
Sbjct: 3 VKLLDLDGSQGHKEWLTEVVFLGQLRHPHLVKLIGYCCEEEHRLLVYEYLPRGSLENQLF 182
Query: 172 ELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFG 231
+ P WSTRMK+A GAA+GL +LH A PVIYRD K++NILLD+++N KLSDFG
Sbjct: 183 RIFTASLP--WSTRMKIAAGAAKGLAFLH-EAKKPVIYRDFKASNILLDSDYNAKLSDFG 353
Query: 232 LAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGK 264
LAK GP GD+THVSTRVMGT GY APEY M+GK
Sbjct: 354 LAKDGPEGDDTHVSTRVMGTQGYAAPEYIMTGK 452
Score = 97.8 bits (242), Expect(2) = 1e-70
Identities = 48/99 (48%), Positives = 65/99 (65%)
Frame = +2
Query: 263 GKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQ 322
G LT SD+YSFGVVLLELLTGRR++D R EQNLV WAR +D R+ ++DP L+
Sbjct: 563 GHLTAMSDVYSFGVVLLELLTGRRSVDKGRPQREQNLVEWARSALNDSRKLSRIMDPRLE 742
Query: 323 GRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
G++ +A A+ CL +P+ RPL++ +V LE L
Sbjct: 743 GQYSEVGARKAAALAYQCLSHRPRSRPLMSTVVNVLEPL 859
>TC230612 similar to GB|BAA98102.1|8978074|AB025609 protein serine/threonine
kinase-like {Arabidopsis thaliana;} , partial (70%)
Length = 1606
Score = 262 bits (669), Expect = 2e-70
Identities = 149/369 (40%), Positives = 215/369 (57%), Gaps = 14/369 (3%)
Frame = +3
Query: 5 SCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGSS 64
+C F + K S E+ + + GN G + ++ ++ R K K
Sbjct: 39 NCFFLKNKSKSDAELPKRRKKK-NQVGNNGASKSSTSSPLPSPRSIKELYK--------- 188
Query: 65 NGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEA-------VAVKQLSH 117
+ F +EL DAT F IGEGGFGKVY+G + VA+K+L+
Sbjct: 189 ENEHNFRIFTLQELVDATHGFNRMLKIGEGGFGKVYRGTIKPHPEDGADPILVAIKKLNT 368
Query: 118 DGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCT----DGDQRLLVYEYMP---MGSLEDHL 170
G QG +E++ EV LS+++H NLV+L+GYC+ G QRLLVYE+M G +
Sbjct: 369 RGLQGHKEWLAEVQFLSVVNHPNLVKLLGYCSVDSEKGIQRLLVYEFMSNRESGKITFFS 548
Query: 171 FELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDF 230
L H L W TR+++ +GAA+GL YLH + VIYRD KS+N+LLD +F+PKLSDF
Sbjct: 549 LSLPH----LTWKTRLQIMLGAAQGLHYLHNGLEVKVIYRDFKSSNVLLDKKFHPKLSDF 716
Query: 231 GLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDT 290
GLA+ GP GD THVST V+GT GY APEY +G L ++SDI+SFGVVL E+LTGRR ++
Sbjct: 717 GLAREGPTGDQTHVSTAVVGTQGYAAPEYVETGHLKIQSDIWSFGVVLYEILTGRRVLNR 896
Query: 291 SRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPL 350
+R GEQ L+ W + Y ++ RF ++DP L+ ++ + + CL++ P+ RP
Sbjct: 897 NRPIGEQKLIEWVKNYPANSSRFSKIIDPRLKNQYSLGAARKVAKLADNCLKKNPEDRPS 1076
Query: 351 ITDIVVALE 359
++ IV +L+
Sbjct: 1077MSQIVESLK 1103
>TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (50%)
Length = 1162
Score = 261 bits (667), Expect = 4e-70
Identities = 131/214 (61%), Positives = 158/214 (73%)
Frame = +2
Query: 150 DGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIY 209
+ DQRLLVYE+MP GSLE+HLF PL WS RMK+A+GAA+GL +LH A P+IY
Sbjct: 2 EDDQRLLVYEFMPRGSLENHLFRRPL---PLPWSIRMKIALGAAKGLAFLHEEAQRPIIY 172
Query: 210 RDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKS 269
RD K++NILLD E+N KLSDFGLAK GP G+ THVSTRVMGTYGY APEY M+G LT KS
Sbjct: 173 RDFKTSNILLDAEYNAKLSDFGLAKDGPEGEKTHVSTRVMGTYGYAAPEYVMTGHLTSKS 352
Query: 270 DIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRC 329
D+YSFGVVLLE+LTGRR+ID R GE NLV WARP DRR F ++DP L+G F +
Sbjct: 353 DVYSFGVVLLEMLTGRRSIDKKRPNGEHNLVEWARPVLGDRRMFYRIIDPRLEGHFSVKG 532
Query: 330 LHQAIAITAMCLQEQPKFRPLITDIVVALEYLAS 363
+A + A CL PK RPL++++V AL+ L S
Sbjct: 533 AQKAALLAAQCLSRDPKSRPLMSEVVRALKPLPS 634
>TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase C, epsilon
type (nPKC-epsilon) , partial (5%)
Length = 908
Score = 260 bits (664), Expect = 8e-70
Identities = 135/256 (52%), Positives = 174/256 (67%), Gaps = 10/256 (3%)
Frame = +3
Query: 77 ELADATRNFKEANLIGEGGFGKVYKG----------RLTTGEAVAVKQLSHDGRQGFQEF 126
EL ATRNF+ ++GEGGFG+V+KG R+ G VAVK+ + D QG +E+
Sbjct: 150 ELRSATRNFRPDTVLGEGGFGRVFKGWIDKNTFKPSRVGVGIPVAVKKSNPDSLQGLEEW 329
Query: 127 VMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRM 186
EV +L H NL +LIGYC + Q LLVYEYM GSLE HLF +PL+W R+
Sbjct: 330 QSEVQLLGKFSHPNLGKLIGYCWEESQFLLVYEYMQKGSLESHLFR--RGPKPLSWDIRL 503
Query: 187 KVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST 246
K+A+GAARGL +LH T++ VIYRD KS+NILLD +FN KLSDFGLAK GPV +HV+T
Sbjct: 504 KIAIGAARGLAFLH-TSEKSVIYRDFKSSNILLDGDFNAKLSDFGLAKFGPVNGKSHVTT 680
Query: 247 RVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPY 306
R+MGTYGY APEY +G L +KSD+Y FGVVLLE+LTGR A+DT++ G QNLV
Sbjct: 681 RIMGTYGYAAPEYMATGHLYIKSDVYGFGVVLLEMLTGRAALDTNQPTGMQNLVECTMSS 860
Query: 307 FSDRRRFGHMVDPLLQ 322
++R ++DP ++
Sbjct: 861 LHAKKRLKEVMDPNME 908
>TC227460 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (44%)
Length = 581
Score = 257 bits (657), Expect = 5e-69
Identities = 128/196 (65%), Positives = 152/196 (77%)
Frame = +3
Query: 130 VLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVA 189
V L L H NLV+L+GYC + DQRLLVYE+MP GSLE+HLF S PL WS RMK+A
Sbjct: 3 VNFLGDLVHPNLVKLVGYCIEEDQRLLVYEFMPRGSLENHLFRRSI---PLPWSIRMKIA 173
Query: 190 VGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVM 249
+GAA+GL +LH A+ PVIYRD K++NILLD E+N KLSDFGLAK GP GD THVSTRVM
Sbjct: 174 LGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFGLAKDGPEGDKTHVSTRVM 353
Query: 250 GTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSD 309
GTYGY APEY M+G LT KSD+YSFGVVLLE+LTGRR++D R GE NLV WA+P+ +
Sbjct: 354 GTYGYAAPEYVMTGHLTSKSDVYSFGVVLLEMLTGRRSMDKHRPNGEHNLVEWAQPHLGE 533
Query: 310 RRRFGHMVDPLLQGRF 325
RRRF ++DP L+ F
Sbjct: 534 RRRFYKLIDPHLESHF 581
>TC229394 similar to GB|BAA98102.1|8978074|AB025609 protein serine/threonine
kinase-like {Arabidopsis thaliana;} , partial (63%)
Length = 1170
Score = 250 bits (638), Expect = 9e-67
Identities = 134/279 (48%), Positives = 190/279 (68%), Gaps = 11/279 (3%)
Frame = +3
Query: 92 GEGGFGKVYKGRLTTGEA------VAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLI 145
GEGGFG VYKG + + VA+K+L+ G QG +E++ EV L +++H NLV+L+
Sbjct: 3 GEGGFGSVYKGSIAQLDGQGDPIPVAIKRLNTRGFQGHKEWLAEVQFLGIVNHPNLVKLL 182
Query: 146 GYCT-DGD---QRLLVYEYMPMGSLEDHLFELSHDKEP-LNWSTRMKVAVGAARGLEYLH 200
GYC+ DG+ QRLLVYE+MP SLEDHLF + K P L W TR+++ +GAA+GL YLH
Sbjct: 183 GYCSVDGERGIQRLLVYEFMPNRSLEDHLF---NKKLPTLPWKTRLEIMLGAAQGLAYLH 353
Query: 201 CTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYA 260
+ VIYRD KS+N+LLD +F+PKLSD GLA+ GP GD THVST V+GT GY APEY
Sbjct: 354 EGLEIQVIYRDFKSSNVLLDADFHPKLSDXGLAREGPQGDQTHVSTAVVGTQGYAAPEYI 533
Query: 261 MSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPL 320
+G L ++SD++SFGVVL E+LTGRR+++ +R EQ L+ W + Y +D RF ++D
Sbjct: 534 ETGHLKVQSDMWSFGVVLYEILTGRRSLERNRPTAEQKLLDWVKQYPADTSRFVIIMDAR 713
Query: 321 LQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
L+ ++ + + CL++ P+ RP ++ IV +L+
Sbjct: 714 LRNQYSLPAARKIAKLADSCLKKNPEDRPSMSQIVESLK 830
>TC226990 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (50%)
Length = 991
Score = 250 bits (638), Expect = 9e-67
Identities = 118/166 (71%), Positives = 143/166 (86%)
Frame = +1
Query: 199 LHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPE 258
LH A+PPVIYRD KS+NILLD ++PKLSDFGLAKLGPVGD +HVSTRVMGTYGYCAPE
Sbjct: 4 LHDKANPPVIYRDFKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPE 183
Query: 259 YAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVD 318
YAM+G+LT+KSD+YSFGVV LEL+TGR+AID++R GEQNLV+WARP FSDRR+F + D
Sbjct: 184 YAMTGQLTVKSDVYSFGVVFLELITGRKAIDSTRPHGEQNLVTWARPLFSDRRKFPKLAD 363
Query: 319 PLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
P LQGR+P R L+QA+A+ +MC+QEQ RPLI D+V AL +LA+Q
Sbjct: 364 PQLQGRYPMRGLYQALAVASMCIQEQAAARPLIGDVVTALSFLANQ 501
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,203,986
Number of Sequences: 63676
Number of extensions: 218585
Number of successful extensions: 2875
Number of sequences better than 10.0: 988
Number of HSP's better than 10.0 without gapping: 2141
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2182
length of query: 385
length of database: 12,639,632
effective HSP length: 99
effective length of query: 286
effective length of database: 6,335,708
effective search space: 1812012488
effective search space used: 1812012488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0193.1


