

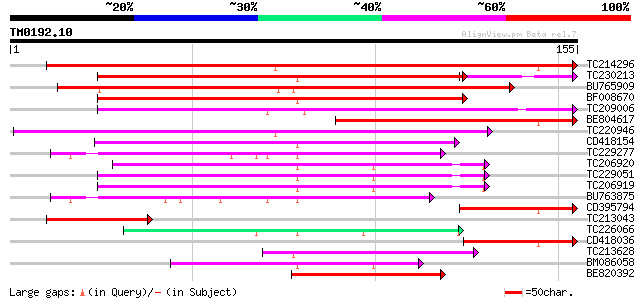
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0192.10
(155 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214296 similar to UP|Q9VH98 (Q9VH98) CG8348-PA, partial (5%) 221 1e-58
TC230213 weakly similar to UP|Q9ZTM8 (Q9ZTM8) PGPS/D12, partial ... 136 8e-36
BU765909 118 1e-27
BF008670 weakly similar to GP|8778226|gb|A F10B6.27 {Arabidopsis... 107 2e-24
TC209006 weakly similar to UP|Q9LS45 (Q9LS45) Arabidopsis thalia... 104 1e-23
BE804617 weakly similar to GP|27414003|gb| fw2.2 {Lycopersicon c... 103 3e-23
TC220946 weakly similar to UP|Q8GRS4 (Q8GRS4) Fw2.2, partial (39%) 98 1e-21
CD418154 weakly similar to GP|13605847|gb| AT4g35920/F4B14_190 {... 82 1e-16
TC229277 69 9e-13
TC206920 weakly similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial ... 60 4e-10
TC229051 similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial (13%) 58 2e-09
TC206919 weakly similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial ... 58 2e-09
BU763875 51 3e-07
CD395794 50 3e-07
TC213043 50 6e-07
TC226066 similar to UP|Q6ULR9 (Q6ULR9) SAT5, partial (80%) 48 2e-06
CD418036 similar to GP|16930765|gb|A photosystem II reaction cen... 48 2e-06
TC213628 46 8e-06
BM086058 43 5e-05
BE820392 43 5e-05
>TC214296 similar to UP|Q9VH98 (Q9VH98) CG8348-PA, partial (5%)
Length = 1491
Score = 221 bits (563), Expect = 1e-58
Identities = 101/152 (66%), Positives = 117/152 (76%), Gaps = 7/152 (4%)
Frame = +2
Query: 11 YQPTPPPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFY 70
Y P PPP P+P V+WSTGL DCFS+ NCC+T WCPCVTFGR+AEIVDKGSTSCGASG
Sbjct: 548 YAPVPPPQPKPLVDWSTGLCDCFSECGNCCMTCWCPCVTFGRVAEIVDKGSTSCGASGAL 727
Query: 71 F------VQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFN 124
+ + G YS YR K+R QY LKGN C DCL HCFC CALCQEYREL+ +GF+
Sbjct: 728 YTLICCVIGCGCLYSCFYRPKMRRQYGLKGNGCSDCLIHCFCEPCALCQEYRELQHRGFD 907
Query: 125 MKIGWHGNVEQQTRGVAMT-STAPTVEQSMTR 155
M IGWHGNVEQ++RGVAMT +TAP+VE M+R
Sbjct: 908 MIIGWHGNVEQRSRGVAMTATTAPSVENGMSR 1003
>TC230213 weakly similar to UP|Q9ZTM8 (Q9ZTM8) PGPS/D12, partial (16%)
Length = 767
Score = 136 bits (342), Expect(2) = 8e-36
Identities = 61/107 (57%), Positives = 75/107 (70%), Gaps = 6/107 (5%)
Frame = +2
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF------Y 78
WS+GL CFSD ++CCLT+WCPC +FGRI EIVDKG+TSC G F G F Y
Sbjct: 149 WSSGLCGCFSDCSSCCLTFWCPCASFGRIGEIVDKGTTSCCLHGSLFCLLGGFSYLAGIY 328
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNM 125
+ YRTKIR QY ++G+ C D L CFCS C LCQEYREL+ +GF++
Sbjct: 329 ACMYRTKIRRQYGIEGHQCADFLLSCFCSACTLCQEYRELQARGFDV 469
Score = 30.4 bits (67), Expect(2) = 8e-36
Identities = 15/32 (46%), Positives = 19/32 (58%)
Frame = +3
Query: 124 NMKIGWHGNVEQQTRGVAMTSTAPTVEQSMTR 155
N IGW GNV+ +T GV + AP V M+R
Sbjct: 477 NAYIGWKGNVQMRTSGV---TAAPVVNGGMSR 563
>BU765909
Length = 421
Score = 118 bits (295), Expect = 1e-27
Identities = 60/130 (46%), Positives = 82/130 (62%), Gaps = 5/130 (3%)
Frame = +1
Query: 14 TPPPPPQPPV--EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYF 71
T P + PV +W+TGLYDC+ D ++CC T++CPC+TFG+IAEIVD G+ S A+ +
Sbjct: 28 TYKPYGKQPVKGQWTTGLYDCWDDPSHCCFTWFCPCITFGQIAEIVDGGTISKNAACCIY 207
Query: 72 V-QHGV--FYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIG 128
V HG Y A YR+K+R ++L D H C CAL QEY+EL+ +G + IG
Sbjct: 208 VDSHGTKWLYGATYRSKLRRLFSLSQEPYSDPFLHGCCCICALTQEYKELKNRGIDPSIG 387
Query: 129 WHGNVEQQTR 138
W GNVE+ R
Sbjct: 388 WEGNVEKWKR 417
>BF008670 weakly similar to GP|8778226|gb|A F10B6.27 {Arabidopsis thaliana},
partial (28%)
Length = 346
Score = 107 bits (268), Expect = 2e-24
Identities = 50/107 (46%), Positives = 67/107 (61%), Gaps = 6/107 (5%)
Frame = +2
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF------Y 78
WS+GL CFSD +CCL++WCPC FGRI EIV G+TSC G F G F Y
Sbjct: 26 WSSGLCGCFSDCCSCCLSFWCPCACFGRIREIVVIGTTSCCLHGSLFCLLGRFSYLAGIY 205
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNM 125
+ +RT I+ Y + G+ C L +CFCS C LC ++REL+ +GF++
Sbjct: 206 ACMFRTLIKKLYRIVGHLCAVSLLNCFCSACTLCLDFRELQARGFDV 346
>TC209006 weakly similar to UP|Q9LS45 (Q9LS45) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MYF24 (At3g18450), partial
(23%)
Length = 792
Score = 104 bits (260), Expect = 1e-23
Identities = 55/137 (40%), Positives = 78/137 (56%), Gaps = 6/137 (4%)
Frame = +1
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGF-YFVQHGVFYS---- 79
WSTGL+DC + N +T + PCVTFG+IAE+ D G SC F Y + S
Sbjct: 97 WSTGLFDCHENQTNAVMTAFFPCVTFGQIAEVQDGGELSCHLGSFIYLLMMPALCSQWIM 276
Query: 80 -ADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQTR 138
+ YRTK+R +YNL D ++H FC C+LCQE+REL+ +G + +GW+G + QQ
Sbjct: 277 GSKYRTKLRKRYNLVEAPYTDIVSHIFCPCCSLCQEFRELKIRGLDPALGWNGILAQQQS 456
Query: 139 GVAMTSTAPTVEQSMTR 155
T P + Q M++
Sbjct: 457 D--QTLKNPPLNQVMSK 501
>BE804617 weakly similar to GP|27414003|gb| fw2.2 {Lycopersicon cheesmanii},
partial (32%)
Length = 404
Score = 103 bits (257), Expect = 3e-23
Identities = 47/67 (70%), Positives = 55/67 (81%), Gaps = 1/67 (1%)
Frame = +2
Query: 90 YNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQTRGVAMT-STAPT 148
Y+L+GN C DCL H FC CALCQEYREL+ +GF+M IGWHGNVEQ++RGVAMT +TAP
Sbjct: 2 YDLRGNACTDCLIHFFCEPCALCQEYRELQFRGFHMTIGWHGNVEQRSRGVAMTVATAPP 181
Query: 149 VEQSMTR 155
VEQ M R
Sbjct: 182 VEQGMNR 202
>TC220946 weakly similar to UP|Q8GRS4 (Q8GRS4) Fw2.2, partial (39%)
Length = 530
Score = 98.2 bits (243), Expect = 1e-21
Identities = 51/136 (37%), Positives = 70/136 (50%), Gaps = 5/136 (3%)
Frame = +2
Query: 2 YKTVAGSTDYQPTPPPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGS 61
+K G P P +W+TGL+DCF D CC T+ CP FG AEI+D+G
Sbjct: 17 FKMDTGEGKSAQAPNINAPPTGQWTTGLFDCFDDTGICCSTWLCPQCIFGPNAEIIDQGR 196
Query: 62 TSCGASGFYF-----VQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYR 116
TS ++ + F V YS +R+K+R+ YNL C D H C A+ QE R
Sbjct: 197 TSSRSATYIFCGLSLVGWAFLYSFKFRSKLRALYNLPEEPCGDLCVHYCCLVFAISQERR 376
Query: 117 ELEKQGFNMKIGWHGN 132
EL+ +G + +GW GN
Sbjct: 377 ELKNRGLDTSVGWKGN 424
>CD418154 weakly similar to GP|13605847|gb| AT4g35920/F4B14_190 {Arabidopsis
thaliana}, partial (19%)
Length = 595
Score = 81.6 bits (200), Expect = 1e-16
Identities = 43/105 (40%), Positives = 58/105 (54%), Gaps = 5/105 (4%)
Frame = -3
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF-----Y 78
EW T L+ C + C T + PC TF IA +V +G TS + V + +F Y
Sbjct: 566 EWKTDLFGCCREPCLCLKTCFFPCGTFSWIANVVTRGETSRKRAMTNLVAYSIFCGCCCY 387
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGF 123
S R K+R+Q+N++G C D LTH C CA+ QE+RELE GF
Sbjct: 386 SCCIRRKLRNQFNIEGGLCDDFLTHLMCCCCAMVQEWRELELSGF 252
>TC229277
Length = 1056
Score = 68.9 bits (167), Expect = 9e-13
Identities = 44/147 (29%), Positives = 64/147 (42%), Gaps = 39/147 (26%)
Frame = +2
Query: 12 QPTP----PPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDK-----GST 62
QP P PP +P W TG++ C D NC +CPCV FGR E + + G
Sbjct: 296 QPLPESYTPPADEP---WMTGIFGCTGDRENCLTGLFCPCVLFGRNVETLHEETPWTGPC 466
Query: 63 SCGA----------------SGF------YFVQHGVF--------YSADYRTKIRSQYNL 92
C A +GF + + G+F Y+ R ++ +Y+L
Sbjct: 467 VCHAIFVEGGIALATATAIFNGFIDPGTSFLIFEGLFFTWWMCGIYTGQVRQNLQKKYHL 646
Query: 93 KGNNCLDCLTHCFCSRCALCQEYRELE 119
+ + C C HC CALCQE+RE++
Sbjct: 647 QNSPCDPCCVHCCMHWCALCQEHREMK 727
>TC206920 weakly similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial (7%)
Length = 970
Score = 60.1 bits (144), Expect = 4e-10
Identities = 40/133 (30%), Positives = 58/133 (43%), Gaps = 30/133 (22%)
Frame = +3
Query: 29 LYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF----------- 77
L DCF D + +CPC FG+ ++ GS A ++ + G F
Sbjct: 18 LLDCFDDRRIAFESAFCPCYRFGKNMKLAGFGSCYIQAIVYFLLAIGAFVTFIAYTITRT 197
Query: 78 ----------------YSADYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQEYRELE 119
Y YRT++R ++N+KG++ DC+ H C C LCQE R LE
Sbjct: 198 HYFLYPAVALIIVVGAYLGFYRTRMRKKFNIKGSDSSLDDCIYHFVCPCCTLCQESRTLE 377
Query: 120 KQGFNMKIG-WHG 131
N++ G WHG
Sbjct: 378 IN--NVQNGTWHG 410
>TC229051 similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial (13%)
Length = 961
Score = 58.2 bits (139), Expect = 2e-09
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 30/137 (21%)
Frame = +1
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF------- 77
W + DCF D+ + CPC FG+ + GS A+ ++ + G F
Sbjct: 259 WEGEVLDCFDDHRIAIESTCCPCYRFGKNMKRAGFGSCYIQAAIYFLLAVGAFLNFIAFA 438
Query: 78 --------------------YSADYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQEY 115
Y +RT++R ++N+ G++ DC+ H C C LCQE
Sbjct: 439 VTRRHCYLYLTVAFVVSVGAYLGFFRTRLRKKFNIMGSDSSMDDCVYHFACPCCTLCQES 618
Query: 116 RELEKQGFNMKIG-WHG 131
R LE N++ G WHG
Sbjct: 619 RTLEMN--NVRDGTWHG 663
>TC206919 weakly similar to UP|Q9LPZ7 (Q9LPZ7) T23J18.4, partial (15%)
Length = 1095
Score = 57.8 bits (138), Expect = 2e-09
Identities = 40/137 (29%), Positives = 57/137 (41%), Gaps = 30/137 (21%)
Frame = +2
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF------- 77
W L DCF D + CPC FG+ ++ GS A ++ + G F
Sbjct: 245 WEGELLDCFDDRRIAFESACCPCYRFGKNMKLAGFGSCYIQAIVYFLLAIGAFVTSIAYT 424
Query: 78 --------------------YSADYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQEY 115
Y YRT++R ++N+KG++ D + H C C LCQE
Sbjct: 425 ITRTHYFLYLAVAFIIAVGAYLGFYRTRMRKKFNIKGSDSSLDDFVYHFVCPCCTLCQES 604
Query: 116 RELEKQGFNMKIG-WHG 131
R LE N++ G WHG
Sbjct: 605 RTLEMN--NVQNGTWHG 649
>BU763875
Length = 438
Score = 50.8 bits (120), Expect = 3e-07
Identities = 37/128 (28%), Positives = 56/128 (42%), Gaps = 23/128 (17%)
Frame = +3
Query: 12 QPTP----PPPPQPPVEWSTGLYDCFSDYNNCCL---TYWC-PCVTFGRIAEI-VDKGST 62
QP P PP +P W TG++ C D N L T W PC+ E + +
Sbjct: 63 QPLPESYTPPADEP---WMTGIFGCTEDREN*TLHEETPWTGPCICHAIFVEGGIALATA 233
Query: 63 SCGASGF------YFVQHGVF--------YSADYRTKIRSQYNLKGNNCLDCLTHCFCSR 108
+ +GF + + G+F Y+ R ++ +Y+L+ + C C HC
Sbjct: 234 TAIFNGFIVPGTSFLIFEGLFFTWWMCGIYTGQVRQNLQKKYHLENSPCDPCCVHCCMHW 413
Query: 109 CALCQEYR 116
CALCQE+R
Sbjct: 414 CALCQEHR 437
>CD395794
Length = 269
Score = 50.4 bits (119), Expect = 3e-07
Identities = 24/33 (72%), Positives = 28/33 (84%), Gaps = 1/33 (3%)
Frame = -2
Query: 124 NMKIGWHGNVEQQTRGVAMT-STAPTVEQSMTR 155
+M IGWHGNVEQ++RGVAMT +TAP VEQ M R
Sbjct: 268 HMTIGWHGNVEQRSRGVAMTVATAPPVEQGMNR 170
>TC213043
Length = 439
Score = 49.7 bits (117), Expect = 6e-07
Identities = 19/29 (65%), Positives = 21/29 (71%)
Frame = +1
Query: 11 YQPTPPPPPQPPVEWSTGLYDCFSDYNNC 39
Y P PPP P+P V WSTGL DCFS+ NC
Sbjct: 136 YAPVPPPQPKPLVNWSTGLCDCFSECGNC 222
>TC226066 similar to UP|Q6ULR9 (Q6ULR9) SAT5, partial (80%)
Length = 1053
Score = 47.8 bits (112), Expect = 2e-06
Identities = 39/135 (28%), Positives = 52/135 (37%), Gaps = 42/135 (31%)
Frame = +2
Query: 32 CFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGA-----SGFYFVQHGVF--------Y 78
C SD C L PCV +G E + + SG Y + + F +
Sbjct: 353 CSSDLEVCLLGSVAPCVLYGSNVERLGSARGTFANHCLPYSGMYIIGNSCFGWNCLAPWF 532
Query: 79 SADYRTKIRSQYNLKGN-------------------------NCLDCLTHCFCSRCALCQ 113
S RT IR ++NL+G+ + D TH FC CALCQ
Sbjct: 533 SYPSRTAIRRRFNLEGSCEALNRSCGCCGSILEDEVQREHCESACDLATHVFCHVCALCQ 712
Query: 114 EYRELEKQ----GFN 124
E REL ++ GFN
Sbjct: 713 EGRELRRRLPHPGFN 757
>CD418036 similar to GP|16930765|gb|A photosystem II reaction center {Retama
raetam}, partial (94%)
Length = 618
Score = 47.8 bits (112), Expect = 2e-06
Identities = 23/32 (71%), Positives = 27/32 (83%), Gaps = 1/32 (3%)
Frame = -1
Query: 125 MKIGWHGNVEQQTRGVAMT-STAPTVEQSMTR 155
M IGWHGNVEQ++RGVAMT +TAP+VE M R
Sbjct: 618 MIIGWHGNVEQRSRGVAMTATTAPSVENGMIR 523
>TC213628
Length = 994
Score = 45.8 bits (107), Expect = 8e-06
Identities = 24/61 (39%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Frame = +1
Query: 70 YFVQHGV--FYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKI 127
Y HG Y A YR+K+R ++L D H C CAL QEY+EL+ +G + I
Sbjct: 232 YVDSHGTKWLYGATYRSKLRRLFSLSQEPYSDPFLHGCCCICALTQEYKELKNRGIDPSI 411
Query: 128 G 128
G
Sbjct: 412 G 414
>BM086058
Length = 407
Score = 43.1 bits (100), Expect = 5e-05
Identities = 26/98 (26%), Positives = 40/98 (40%), Gaps = 29/98 (29%)
Frame = +3
Query: 45 CPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF--------------------------- 77
CPC FG+ ++ GS A ++ + G F
Sbjct: 33 CPCYRFGKNMKLAGFGSCYIQAIVYFLLAIGAFVTFIAYTITRTHYFLYPAVALIIVVGA 212
Query: 78 YSADYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQ 113
Y YRT++R ++N+KG++ DC+ H C C LCQ
Sbjct: 213 YLGFYRTRMRKKFNIKGSDSSLDDCIYHFVCPCCTLCQ 326
>BE820392
Length = 685
Score = 43.1 bits (100), Expect = 5e-05
Identities = 16/42 (38%), Positives = 26/42 (61%)
Frame = -3
Query: 78 YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELE 119
Y+ R ++ +Y+L+ + C C HC CALCQE+RE++
Sbjct: 398 YTGQVRQNLQKKYHLENSPCDPCCVHCCMHWCALCQEHREMK 273
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,157,248
Number of Sequences: 63676
Number of extensions: 251828
Number of successful extensions: 7748
Number of sequences better than 10.0: 264
Number of HSP's better than 10.0 without gapping: 3668
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6401
length of query: 155
length of database: 12,639,632
effective HSP length: 89
effective length of query: 66
effective length of database: 6,972,468
effective search space: 460182888
effective search space used: 460182888
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0192.10


