

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0191.8
(1758 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
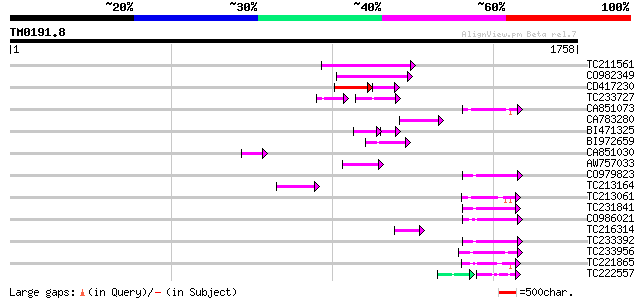
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 174 2e-43
CO982349 138 2e-32
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 88 1e-24
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 74 7e-23
CA851073 85 2e-16
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 84 7e-16
BI471325 64 3e-15
BI972659 79 1e-14
CA851030 78 4e-14
AW757033 77 5e-14
CO979823 77 5e-14
TC213164 75 3e-13
TC213061 74 7e-13
TC231841 72 2e-12
CO986021 72 2e-12
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 72 3e-12
TC233392 71 4e-12
TC233956 70 8e-12
TC221865 69 1e-11
TC222557 53 2e-11
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase (Fragment)
, partial (59%)
Length = 1077
Score = 174 bits (442), Expect = 2e-43
Identities = 95/290 (32%), Positives = 146/290 (49%)
Frame = +1
Query: 968 RKGKKGYMALKLDMSKAYDRIEWDFLKQVLTTMGFPSKITELISICVSSVSYSLLVNGEP 1027
++ K + K+D KAYD + W+FL ++ M F K + I CV S S S+LVNG P
Sbjct: 43 KRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEECVKSASISVLVNGSP 222
Query: 1028 SPPFSPQRGLRQGDPLSPYLFILCVEAFSGLISNQIRLRNLHGIKVARGAPPINHLFFAD 1087
+ F PQRGLRQGDPL+P+LF + E +GL+ + V PI+ L +AD
Sbjct: 223 TAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLVGANGVPISILQYAD 402
Query: 1088 DSLIFSRATQEEASSLLDVIRQYEQASGQLVNMDKSEISFSRNVPQTSMDTIRNRMGVKA 1147
D++ F A +E ++ ++R +E S +N KS Q + N +
Sbjct: 403 DTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGVTDQWKQEA-ANYLHCSL 579
Query: 1148 VIVHDKYLGLPTVIGHSKKAIFGKIQERVWRKLKGWKEKTLSKAGKEILIKAVAQAIPSY 1207
+ YLG+P + ++ I + RKL WK + +S G+ LIK+V +IP Y
Sbjct: 580 LAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGRVTLIKSVLTSIPIY 759
Query: 1208 VMSCFKLPNGICSKIESLLANFWWGQKENERKIHWAAWQHLCKTKNRGDL 1257
S F++P + K+ + F WG +++KI W W+ LC K RG +
Sbjct: 760 FFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKERGGI 909
>CO982349
Length = 795
Score = 138 bits (348), Expect = 2e-32
Identities = 75/237 (31%), Positives = 118/237 (49%)
Frame = +3
Query: 1013 CVSSVSYSLLVNGEPSPPFSPQRGLRQGDPLSPYLFILCVEAFSGLISNQIRLRNLHGIK 1072
C+ S S S+LVN P FSPQRGLRQGDPL+P LF + E +GL+ + + +
Sbjct: 81 CLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRFNSFL 260
Query: 1073 VARGAPPINHLFFADDSLIFSRATQEEASSLLDVIRQYEQASGQLVNMDKSEISFSRNVP 1132
V + P++ L +ADD++ F AT E + ++R +E ASG +N +S P
Sbjct: 261 VGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGAIWK-P 437
Query: 1133 QTSMDTIRNRMGVKAVIVHDKYLGLPTVIGHSKKAIFGKIQERVWRKLKGWKEKTLSKAG 1192
+ + + YLG+P + I I + KL WK++ +S G
Sbjct: 438 DHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHISLGG 617
Query: 1193 KEILIKAVAQAIPSYVMSCFKLPNGICSKIESLLANFWWGQKENERKIHWAAWQHLC 1249
+ LI A+ A+P Y S F+ P+ + K+ ++ F WG +R+I W W+ +C
Sbjct: 618 RVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWVKWETVC 788
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like protein
{Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 87.8 bits (216), Expect(2) = 1e-24
Identities = 45/117 (38%), Positives = 71/117 (60%)
Frame = -3
Query: 1008 ELISICVSSVSYSLLVNGEPSPPFSPQRGLRQGDPLSPYLFILCVEAFSGLISNQIRLRN 1067
+L+ C+S+ + SLL NGE FSP G+RQ DP++PYLF+LC+E S LI+ + +
Sbjct: 683 QLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL 504
Query: 1068 LHGIKVARGAPPINHLFFADDSLIFSRATQEEASSLLDVIRQYEQASGQLVNMDKSE 1124
I+V RG I+HL F DD ++F A ++ + + + +SG VN+DK++
Sbjct: 503 *RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTK 333
Score = 45.4 bits (106), Expect(2) = 1e-24
Identities = 27/86 (31%), Positives = 45/86 (51%)
Frame = -2
Query: 1124 EISFSRNVPQTSMDTIRNRMGVKAVIVHDKYLGLPTVIGHSKKAIFGKIQERVWRKLKGW 1183
+ SFS+NV + I + +G+++ KYLG+P + F I ++V ++ W
Sbjct: 333 KFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRW 154
Query: 1184 KEKTLSKAGKEILIKAVAQAIPSYVM 1209
K LS G+ L K V A+PS++M
Sbjct: 153 KSNLLSMVGRLTLTK*VV*ALPSHIM 76
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 73.6 bits (179), Expect(2) = 7e-23
Identities = 43/99 (43%), Positives = 57/99 (57%)
Frame = -3
Query: 950 ITDNALTGFEVFHYMKKKRKGKKGYMALKLDMSKAYDRIEWDFLKQVLTTMGFPSKITEL 1009
I D EV + + KK G G +ALK+D+ KA+D ++ DFL VL G+
Sbjct: 834 IKDCTCVTSEVINMLDKKVFG--GNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNW 661
Query: 1010 ISICVSSVSYSLLVNGEPSPPFSPQRGLRQGDPLSPYLF 1048
I + + S S+ VNGEP FS QRG+RQGDPLSP L+
Sbjct: 660 IRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 53.9 bits (128), Expect(2) = 7e-23
Identities = 34/140 (24%), Positives = 66/140 (46%)
Frame = -2
Query: 1073 VARGAPPINHLFFADDSLIFSRATQEEASSLLDVIRQYEQASGQLVNMDKSEISFSRNVP 1132
V G+ +H+ ADD +IF + T +L++ + Y+ + GQ+++ KS+I F ++P
Sbjct: 475 VPMGSLTRSHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKI-FIGSIP 299
Query: 1133 QTSMDTIRNRMGVKAVIVHDKYLGLPTVIGHSKKAIFGKIQERVWRKLKGWKEKTLSKAG 1192
+ I + + + Y G+P G + + +++ KL WK LS G
Sbjct: 298 GQRLRVISDLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMG 119
Query: 1193 KEILIKAVAQAIPSYVMSCF 1212
+ L+ +V + Y S +
Sbjct: 118 RIQLVNSVIHGMLLYSFSVY 59
>CA851073
Length = 633
Score = 85.1 bits (209), Expect = 2e-16
Identities = 58/200 (29%), Positives = 92/200 (46%), Gaps = 15/200 (7%)
Frame = +1
Query: 1404 EDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFMYR 1463
+D ++W+ G++S KSAY +I T + S + +KT+W++KI + F +R
Sbjct: 25 QDTMVWKADPCGVYSTKSAY------RILMTCNRHVSEANIFKTIWKLKIPPRAAVFSWR 186
Query: 1464 VINNLTPCRANLARRGIQIQE-QCALCSQDVVETQDHLFLQCSWARAAW-----FHHSLG 1517
+I + P R NL RR + IQE +C LC + E DHLF C R W ++ +G
Sbjct: 187 LIKDRLPTRHNLLRRNVSIQENECPLCGYE-QEEADHLFFNCKMTRGLWWESMRWNQIVG 363
Query: 1518 LRMDLVQTSFLEWLQSFCANQEDEIVAQVFHGTW---------AIWKARNESLFNNVQVH 1568
+ F+++ F A + H W +IWK RN +F Q
Sbjct: 364 PLSVSPASHFVQFCDGFGAGRN--------HTRWCGWWIALTSSIWKHRNLLIFQGNQFE 519
Query: 1569 PATAMEKAQNLLAEWKEAQE 1588
P+ M+ A L W + +E
Sbjct: 520 PSKVMDDALFLAWSWLKVRE 579
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase homolog
T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 83.6 bits (205), Expect = 7e-16
Identities = 45/139 (32%), Positives = 69/139 (49%), Gaps = 5/139 (3%)
Frame = +2
Query: 1210 SCFKLPNGICSKIESLLANFWWGQKENERKIHWAAWQHLCKTKNRGDLGFRNLEDFNRAL 1269
S F LP I +K+ESL F WG + + RKI W W+ +C K +G LG ++L FN L
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTL 184
Query: 1270 LAKQAWRLLNSEKSLLYKVWKAKYFPKCDLLKAGVGYQPSYAWRSVWGTLKF-----IEE 1324
L K W L ++ KV ++KY L + G + S W+ + T + ++
Sbjct: 185 LGKWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLKR 364
Query: 1325 GCKWSIGNGKKVSIWKDPW 1343
W +G G ++ W+D W
Sbjct: 365 ETIWKVGGGDRIKFWEDLW 421
>BI471325
Length = 421
Score = 63.5 bits (153), Expect(2) = 3e-15
Identities = 32/85 (37%), Positives = 50/85 (58%)
Frame = +2
Query: 1067 NLHGIKVARGAPPINHLFFADDSLIFSRATQEEASSLLDVIRQYEQASGQLVNMDKSEIS 1126
++H +KV RG ++HL D +F R +EA L +++ +E +SG +VN+ KSEI
Sbjct: 5 DIHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEIC 184
Query: 1127 FSRNVPQTSMDTIRNRMGVKAVIVH 1151
FSRN Q + D+I +G+ I H
Sbjct: 185 FSRNTSQIARDSIFFHLGMSIWICH 259
Score = 38.1 bits (87), Expect(2) = 3e-15
Identities = 22/63 (34%), Positives = 37/63 (57%), Gaps = 2/63 (3%)
Frame = +1
Query: 1150 VHDKYLGLPTVIGHSKKAIFGKIQERVWRKLKGWKEKTLSKAGKE--ILIKAVAQAIPSY 1207
+ D+YL +P++I KKAIF +++ R ++ K +E ++IK VAQ+I +Y
Sbjct: 232 LRDEYLDMPSMIRTKKKAIFNYLRDEFGRIYSTLVQQESFKGQREKCLIIKYVAQSITTY 411
Query: 1208 VMS 1210
MS
Sbjct: 412 CMS 420
>BI972659
Length = 453
Score = 79.3 bits (194), Expect = 1e-14
Identities = 42/140 (30%), Positives = 71/140 (50%)
Frame = +1
Query: 1103 LLDVIRQYEQASGQLVNMDKSEISFSRNVPQTSMDTIRNRMGVKAVIVHDKYLGLPTVIG 1162
L ++R +E SG +N KS+ P + D + + + + + YLG+P +
Sbjct: 34 LKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWAHDAAQ-LLNCRQLDIPFHYLGMPIAVK 210
Query: 1163 HSKKAIFGKIQERVWRKLKGWKEKTLSKAGKEILIKAVAQAIPSYVMSCFKLPNGICSKI 1222
S + ++ + + KL W +K LS G+ LIK+V A+P Y++S FK+P I K+
Sbjct: 211 ASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKIPQRIVDKL 390
Query: 1223 ESLLANFWWGQKENERKIHW 1242
SL F WG ++ +I W
Sbjct: 391 VSLQRTFMWGGNQHHNRISW 450
>CA851030
Length = 358
Score = 77.8 bits (190), Expect = 4e-14
Identities = 36/79 (45%), Positives = 47/79 (58%)
Frame = +1
Query: 719 IMRRKEMENEFAELMGREELYWRQRSRATWLKEGDKNTSFFHGKANQRQKQNRIARIRDD 778
I K +E L+ +EE YW+QRSRA W KE DKNT FFH K QR+ N I +I+DD
Sbjct: 121 IASTKVVEESLDSLLKQEEEYWQQRSRAIWFKERDKNTEFFHRKTLQRKASNSITKIQDD 300
Query: 779 HGNWVYKQDQVTTAFSNYF 797
GN +K DQ+ ++
Sbjct: 301 EGNVCFKPDQIEDILIKFY 357
>AW757033
Length = 441
Score = 77.4 bits (189), Expect = 5e-14
Identities = 43/128 (33%), Positives = 65/128 (50%)
Frame = -2
Query: 1031 FSPQRGLRQGDPLSPYLFILCVEAFSGLISNQIRLRNLHGIKVARGAPPINHLFFADDSL 1090
FSP RGLRQGDPL+P LF + E +GL+ + + V + P++ L +ADD++
Sbjct: 431 FSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADDTI 252
Query: 1091 IFSRATQEEASSLLDVIRQYEQASGQLVNMDKSEISFSRNVPQTSMDTIRNRMGVKAVIV 1150
F AT E + ++R +E ASG +N KS + +VP M + +
Sbjct: 251 FFGEATMENVRVIKTILRGFELASGLKINFAKSRFG-AISVPD*WRKEAAEFMNCSLLSL 75
Query: 1151 HDKYLGLP 1158
YLG+P
Sbjct: 74 PFSYLGIP 51
>CO979823
Length = 853
Score = 77.4 bits (189), Expect = 5e-14
Identities = 52/193 (26%), Positives = 91/193 (46%), Gaps = 8/193 (4%)
Frame = -2
Query: 1404 EDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFMYR 1463
EDK +W+ G +S KS Y+ + + + +W++KI K F +R
Sbjct: 705 EDKWLWKPEPGGHYSTKSGYHVLWGELTEEIQDAD------FAEIWKLKIPTKAAVFAWR 544
Query: 1464 VINNLTPCRANLARRGIQIQEQ-CALCSQDVVETQDHLFLQCSWARAAWFHHS--LGLRM 1520
++ + P ++NL RR + +Q+ C LC+ ++ E HLF C+ W+ + L+
Sbjct: 543 LVRDRLPTKSNLRRRQVMVQDMVCPLCN-NIEEGAAHLFFNCTKTLPLWWESMSWVNLKT 367
Query: 1521 DLVQTSFLEWLQSFCANQEDEIVAQVFHG-----TWAIWKARNESLFNNVQVHPATAMEK 1575
+ QT +LQ + + D + ++ + TW IW+ RN+ +F N H +E
Sbjct: 366 AMPQTPRQHFLQ-YGTDIADGLKSKRWKCWWIALTWTIWQHRNKVVFQNATFHGNKVLED 190
Query: 1576 AQNLLAEWKEAQE 1588
A LL W +A E
Sbjct: 189 ALLLLWSWFKALE 151
>TC213164
Length = 446
Score = 75.1 bits (183), Expect = 3e-13
Identities = 44/133 (33%), Positives = 67/133 (50%)
Frame = +3
Query: 828 LEEEFTADEVAYALNQMHPLKAPGPDGFPALFYQKYWKLVGGDVTSQVLEILNKGVDPAS 887
L F EV + K+PGPDG F +++W ++ D+ + E GV
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 888 LNQTFICLIPKVKAPENPMEFRPISLCNVIMKLVTKCIANRIKDVLGEVVGEQQSAFIPG 947
N F+ LIPKV P + E++PISL I K+V K ++ R K VL ++ E+Q+ F+ G
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 948 RLITDNALTGFEV 960
R N + E+
Sbjct: 378 RHTLHNVVIANEI 416
>TC213061
Length = 823
Score = 73.6 bits (179), Expect = 7e-13
Identities = 52/202 (25%), Positives = 84/202 (40%), Gaps = 20/202 (9%)
Frame = +2
Query: 1402 DSEDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFM 1461
D ED+ +W G + +SAY ++ +K LW++K+ KV F
Sbjct: 119 DIEDQWVWAAESSGSYLARSAY------RVIREGIPEEEQDREFKELWKLKVPMKVTMFA 280
Query: 1462 YRVINNLTPCRANLARRGIQIQEQ-CALCSQDVVETQDHLFLQCSWARAAWFHHSLGLRM 1520
+R++ ++ P R NL R+ +++ E C LC + + E+ HLF CS W+
Sbjct: 281 WRLLKDILPTRDNLRRKRVELHEYVCPLC-RSMDESASHLFFHCSKILPIWWES------ 439
Query: 1521 DLVQTSFLEWLQSFCA---------NQEDEIVAQVFHG----------TWAIWKARNESL 1561
L W++ A +Q V Q G TW IWK RN+ +
Sbjct: 440 -------LSWVKLVGAFPHHPRHHFHQHSHEVYQGLQGNRWKWWWLALTWTIWKHRNDII 598
Query: 1562 FNNVQVHPATAMEKAQNLLAEW 1583
F+N + M+ A L+ W
Sbjct: 599 FSNATFNAHKVMDDAVFLIWTW 664
>TC231841
Length = 791
Score = 72.4 bits (176), Expect = 2e-12
Identities = 48/187 (25%), Positives = 88/187 (46%), Gaps = 8/187 (4%)
Frame = +3
Query: 1405 DKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFMYRV 1464
D+ +W+ G ++ KSAY + + G ++ LW++K+ +K+ F +R+
Sbjct: 63 DQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDGV------FEELWKLKLPSKITIFAWRL 224
Query: 1465 INNLTPCRANLARRGIQIQE-QCALCSQDVVETQDHLFLQCS------WARAAWFHHSLG 1517
I + P R+NL R+ I++ + +C C + E+ HLF CS W +W + LG
Sbjct: 225 IRDRLPTRSNLRRKQIEVDDPRCPFC-RSAEESAAHLFFHCSRIAPVWWESLSWV-NLLG 398
Query: 1518 LRMDLVQTSFLEWLQSFCANQEDEIVAQVFHG-TWAIWKARNESLFNNVQVHPATAMEKA 1576
+ + + FL+ + A + TW IWK RN +F+N + +++A
Sbjct: 399 VFPNHPRQHFLQHIYGVTAGMRASRWKWWWLALTWTIWKQRNNMIFSNGTFNANKILDEA 578
Query: 1577 QNLLAEW 1583
L+ W
Sbjct: 579 IFLIWTW 599
>CO986021
Length = 900
Score = 72.0 bits (175), Expect = 2e-12
Identities = 53/192 (27%), Positives = 84/192 (43%), Gaps = 7/192 (3%)
Frame = -1
Query: 1404 EDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFMYR 1463
+D ++W+ G++S KSAY + + G +S +F K LW++KI + + F +R
Sbjct: 786 QDSMLWKADPTGIYSTKSAYRLL----LPNNRPGQHSSNF--KILWKLKIPPRAELFSWR 625
Query: 1464 VINNLTPCRANLARRGIQIQE-QCALCSQDVVETQDHLFLQCSWARAAWFH-----HSLG 1517
+ + RANL RR + +Q+ C LC E HLF C W+ +LG
Sbjct: 624 LFRDRLSIRANLLRRHVALQDIMCPLCGNH-QEEAGHLFFHCRMTIGLWWESMNWTRTLG 448
Query: 1518 LRMDLVQTSFLEWLQSFCANQEDEIVAQVFHG-TWAIWKARNESLFNNVQVHPATAMEKA 1576
+D F+++ F A + + T IW+ RN LF P M+ A
Sbjct: 447 AFLDDPAAHFIQFSDGFGAQRNHNRRCLWWIALTSTIWQHRNSLLFKGTPFQPPKVMDDA 268
Query: 1577 QNLLAEWKEAQE 1588
W +A E
Sbjct: 267 LFHAWTWLKATE 232
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 71.6 bits (174), Expect = 3e-12
Identities = 33/93 (35%), Positives = 50/93 (53%)
Frame = +2
Query: 1192 GKEILIKAVAQAIPSYVMSCFKLPNGICSKIESLLANFWWGQKENERKIHWAAWQHLCKT 1251
G+ LIK+V A+P +S FK+P I K+ +L F WG ++ +I W W +C
Sbjct: 8 GRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICNP 187
Query: 1252 KNRGDLGFRNLEDFNRALLAKQAWRLLNSEKSL 1284
K G LG ++L +FN AL + W L ++ L
Sbjct: 188 KIDGGLGIKDLSNFNAALRGRWIWGLASNHNQL 286
>TC233392
Length = 1145
Score = 71.2 bits (173), Expect = 4e-12
Identities = 48/190 (25%), Positives = 86/190 (45%), Gaps = 6/190 (3%)
Frame = -1
Query: 1405 DKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHFMYRV 1464
D +W+ + ++S +SAY +Q G+ + LW++KI K F +R+
Sbjct: 656 DTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGA------LQDLWKLKIPAKASIFAWRL 495
Query: 1465 INNLTPCRANLARRGIQIQEQCALCSQDVVETQDHLFLQCSWARAAWFH-----HSLGLR 1519
I + P ++NL RR + +++ + E H+FL+C+ R W+ SLG+
Sbjct: 494 IRDRLPTKSNLHRRQVVLEDSLCPFCRIREEDASHIFLECNKIRPLWWESQTWVRSLGVS 315
Query: 1520 MDLVQTSFLEWLQSFCANQE-DEIVAQVFHGTWAIWKARNESLFNNVQVHPATAMEKAQN 1578
+ FL+ + ++ + TW++WK RN+ +F N Q + +E A
Sbjct: 314 PINKRQHFLQHVHGTPGSKRYNRWKTWWIALTWSLWKHRNQVIFQNAQFNGIKLLEDAVF 135
Query: 1579 LLAEWKEAQE 1588
L W A E
Sbjct: 134 LHWTWIRAME 105
>TC233956
Length = 757
Score = 70.1 bits (170), Expect = 8e-12
Identities = 55/210 (26%), Positives = 87/210 (41%), Gaps = 12/210 (5%)
Frame = +2
Query: 1393 IKSIRLSWRDSEDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVK 1452
I IRL+ + D +WR G+ S KSAY I+S+ +K LW +K
Sbjct: 26 IHEIRLN-NNFNDTWVWRAESTGIISTKSAYQVIKSE------LDDEGQHLGFKKLWEIK 184
Query: 1453 IQNKVKHFMYRVINNLTPCRANLARRGIQIQEQ-CALCSQDVVETQDHLFLQCSWARAAW 1511
+ K F++R++ + P + NL +R IQ+++ C C ET HLF C W
Sbjct: 185 VPPKALSFVWRLLWDRLPTKDNLIKRQIQVEDDLCPFCHSQ-SETASHLFFTCGKIMPLW 361
Query: 1512 FHHSLGLRMDLV-----QTSFLEWLQSFCANQEDEIVAQVFHGTW------AIWKARNES 1560
+ ++ D V +FL+ S + V+ W +IW+ RN+
Sbjct: 362 WEFLSWVKEDKVFHCRPMDNFLQHYSSAASK-----VSNTRRTMWWIAVTNSIWRLRNDI 526
Query: 1561 LFNNVQVHPATAMEKAQNLLAEWKEAQEHH 1590
+F N V + LL W E +
Sbjct: 527 IFQNQTVDITRLTDSTLFLLWTWLRGWERN 616
>TC221865
Length = 1045
Score = 69.3 bits (168), Expect = 1e-11
Identities = 48/199 (24%), Positives = 85/199 (42%), Gaps = 16/199 (8%)
Frame = -3
Query: 1401 RDSEDKLIWRWSRDGMFSVKSAYYQIQSQKIAATASGSNSPSFSWKTLWRVKIQNKVKHF 1460
R D +W+ +G +S +SAY+ +Q + A + P+F + +W++KI K F
Sbjct: 680 RHLTDCWVWKSEPNGHYSTRSAYHLMQHE----AAKANTDPAF--EEIWKLKIPAKAAIF 519
Query: 1461 MYRVINNLTPCRANLARRGIQIQEQCALCSQDVVETQDHLFLQCSWARAAWFHHSLGLRM 1520
R++ + P + NL RR +Q+ L ++ E HLF C+ + W+
Sbjct: 518 A*RLVKDRLPTKNNLRRRQVQLNGTLCLFCRNYEEEASHLFFSCTKTQPVWWE------- 360
Query: 1521 DLVQTSFLEWLQSFCANQEDEIVAQVFHG----------------TWAIWKARNESLFNN 1564
S++ +F AN + F T+ IW+ RN ++F+N
Sbjct: 359 ---SLSWINTSGAFSANPRHHFLQHSFGLLGLKQYNR*KCWWVALTFTIWQHRNRTIFSN 189
Query: 1565 VQVHPATAMEKAQNLLAEW 1583
+ + ME A L+ W
Sbjct: 188 EPFNGSKMMEDAMFLVWSW 132
>TC222557
Length = 1002
Score = 53.1 bits (126), Expect(2) = 2e-11
Identities = 42/148 (28%), Positives = 67/148 (44%), Gaps = 12/148 (8%)
Frame = +1
Query: 1448 LWRVKIQNKVKHFMYRVINNLTPCRANLARRGIQIQE-QCALCSQDVVETQDHLFLQCS- 1505
LW++KI K F +R+I P + NL RR +Q+ C C++ E HLF C
Sbjct: 439 LWQLKIPLKATTFAWRLIKERIPTKGNLWRRRVQLNNLMCPFCNRQEEEAS-HLFFNCPR 615
Query: 1506 -----WARAAWFHHSLGL-----RMDLVQTSFLEWLQSFCANQEDEIVAQVFHGTWAIWK 1555
W AW ++G+ R + +Q + + + A + VA TW+IW+
Sbjct: 616 ILPLWWESLAWTK-TVGVFSPIPRQNYMQHTLISKGKHLQARWKCWWVAL----TWSIWR 780
Query: 1556 ARNESLFNNVQVHPATAMEKAQNLLAEW 1583
RN +F N + + M+ A L+ W
Sbjct: 781 HRNRIVFLNKAFNGSKLMDDAIFLVWSW 864
Score = 35.4 bits (80), Expect(2) = 2e-11
Identities = 31/127 (24%), Positives = 50/127 (38%), Gaps = 13/127 (10%)
Frame = +3
Query: 1328 WSIGNGKKVSIWKDPWLPGPD---SGYVRTQDVPEFAHEKV-----SDLRLWEGGSWNIS 1379
W +G G K+ W+D WL + + Y R ++ + + +WE W +
Sbjct: 69 WKVGCGDKIRFWEDCWLMEQEPLRAKYPRLYNISCQQQKLILVMGFHSANVWE---WKLE 239
Query: 1380 KIDQLF-NPAEAQLIKSIRLSW----RDSEDKLIWRWSRDGMFSVKSAYYQIQSQKIAAT 1434
LF N +A +SW R + D +W+ +G FS +SAY + T
Sbjct: 240 WRRHLFDNEVQAAASFLDDISWGHIDRWTLDCWVWKPEPNGQFSTRSAYRMLLEGAADQT 419
Query: 1435 ASGSNSP 1441
G P
Sbjct: 420 G*GFRGP 440
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.329 0.141 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,283,488
Number of Sequences: 63676
Number of extensions: 1408238
Number of successful extensions: 9564
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 9301
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9537
length of query: 1758
length of database: 12,639,632
effective HSP length: 111
effective length of query: 1647
effective length of database: 5,571,596
effective search space: 9176418612
effective search space used: 9176418612
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0191.8


