

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0191.7
(680 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
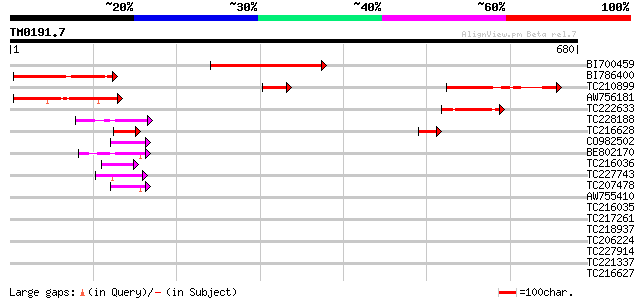
Score E
Sequences producing significant alignments: (bits) Value
BI700459 247 1e-65
BI786400 150 2e-36
TC210899 similar to UP|Q9P115 (Q9P115) Transcription factor ICBP... 147 2e-35
AW756181 122 5e-28
TC222633 79 5e-15
TC228188 similar to GB|AAP31929.1|30387527|BT006585 At3g58030 {A... 46 6e-05
TC216628 weakly similar to UP|Q8H0X2 (Q8H0X2) Expressed protein ... 44 2e-04
CO982502 44 2e-04
BE802170 44 2e-04
TC216036 similar to GB|AAP13393.1|30023720|BT006285 At4g27470 {A... 44 3e-04
TC227743 similar to PIR|E86326|E86326 protein F18O14.3 [imported... 43 4e-04
TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing pr... 42 7e-04
AW755410 42 0.001
TC216035 similar to GB|AAP13393.1|30023720|BT006285 At4g27470 {A... 41 0.002
TC217261 41 0.002
TC218937 similar to UP|PEXA_ARATH (Q9SYU4) Peroxisome assembly p... 41 0.002
TC206224 similar to PIR|E86326|E86326 protein F18O14.3 [imported... 40 0.003
TC227914 similar to UP|Q6R567 (Q6R567) Ring domain containing pr... 40 0.003
TC221337 similar to UP|Q9SD57 (Q9SD57) RNA-binding protein-like ... 40 0.003
TC216627 similar to UP|Q8H0X2 (Q8H0X2) Expressed protein (At1g18... 40 0.004
>BI700459
Length = 423
Score = 247 bits (631), Expect = 1e-65
Identities = 113/140 (80%), Positives = 123/140 (87%)
Frame = +2
Query: 241 PPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVALSG 300
P DHFGPI AENDP RNQG+LVGE+W DR+ECRQWGAH V GIAGQS GAQSV LSG
Sbjct: 2 PTDHFGPITAENDPLRNQGLLVGESWRDRLECRQWGAHFVPVGGIAGQSDRGAQSVVLSG 181
Query: 301 GYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRS 360
GY+DDEDHGEWFLYTGSGG+DLSGNKRTNK SFDQKFE N AL++SC +GYPVRVVRS
Sbjct: 182 GYVDDEDHGEWFLYTGSGGKDLSGNKRTNKSHSFDQKFEKYNRALQVSCLQGYPVRVVRS 361
Query: 361 HKEKRSSYAPESGVRYDGVY 380
HKEKRSSYAPE+GVRYDG+Y
Sbjct: 362 HKEKRSSYAPETGVRYDGIY 421
>BI786400
Length = 367
Score = 150 bits (378), Expect = 2e-36
Identities = 75/127 (59%), Positives = 86/127 (67%), Gaps = 2/127 (1%)
Frame = +3
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCLPTPPASF--VNWDCPDCSSSISDF 62
LPCD+DG CMLCKLKP PS+TL C TC TPWHLPCLP+PP S +WDCPDCS +
Sbjct: 6 LPCDSDGVCMLCKLKPSPSQTLSCGTCATPWHLPCLPSPPLSLSDSHWDCPDCSDTFC-H 182
Query: 63 HAPAPAPSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILDIFDG 122
H AP A LVSAI AI +D SLTDQQKA K+Q+L+ G + K DIFDG
Sbjct: 183 HPVAPT----AHLVSAIHAIQADTSLTDQQKAIKRQQLLAGSAHPSKDKTKA--KDIFDG 344
Query: 123 SINCSFC 129
S+NCS C
Sbjct: 345 SLNCSIC 365
>TC210899 similar to UP|Q9P115 (Q9P115) Transcription factor ICBP90, partial
(3%)
Length = 682
Score = 147 bits (370), Expect = 2e-35
Identities = 83/137 (60%), Positives = 92/137 (66%)
Frame = +3
Query: 525 KSFIRKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIAQDEN 584
+SFIR RAC GGR+LRAQKNVMKCPSCSNDIADFLQNPQVNREM+ V+ESLQ Q
Sbjct: 102 QSFIRNRACGGGRTLRAQKNVMKCPSCSNDIADFLQNPQVNREMLAVIESLQRQAE---- 269
Query: 585 SEQVENSEELSDKSDEIVKNVPDEVEVSKPCDDSTESDENVKNVTDEVEVLKSCDSTEKV 644
ENSEELSDK+DE N+ D EVSKP DST+KV
Sbjct: 270 ----ENSEELSDKNDE---NLHDPTEVSKP-----------------------SDSTDKV 359
Query: 645 LEEINDNDLNRPHKRRK 661
LEEI DN+LN+PHKRRK
Sbjct: 360 LEEIKDNELNQPHKRRK 410
Score = 73.2 bits (178), Expect = 4e-13
Identities = 32/35 (91%), Positives = 32/35 (91%)
Frame = +3
Query: 304 DDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKF 338
DDEDHGEWFLYTGSGGR SGNKRTNKLQSFDQ F
Sbjct: 6 DDEDHGEWFLYTGSGGRXXSGNKRTNKLQSFDQSF 110
Score = 32.3 bits (72), Expect = 0.71
Identities = 15/59 (25%), Positives = 28/59 (47%)
Frame = +3
Query: 52 CPDCSSSISDFHAPAPAPSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGEKP 110
CP CS+ I+DF P ++++ I ++ A ++ + K E + P+ KP
Sbjct: 171 CPSCSNDIADF---LQNPQVNREMLAVIESLQRQAEENSEELSDKNDENLHDPTEVSKP 338
>AW756181
Length = 435
Score = 122 bits (306), Expect = 5e-28
Identities = 64/139 (46%), Positives = 85/139 (61%), Gaps = 8/139 (5%)
Frame = +2
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCLPTPP---ASFVNWDCPDCSSSISD 61
LPCD++G CM+CK P E L C TC TPWH+PCL PP ++ W CPDCS SD
Sbjct: 23 LPCDSEGACMVCKGLPGQEERLLCVTCDTPWHVPCLFAPPPTLSATARWLCPDCSILDSD 202
Query: 62 FHAPAPAPSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPS-----SGEKPPAAGGI 116
P PAP A LV+A+ A+++DASLT KA+K+QEL+ G + ++ +
Sbjct: 203 V-PPVPAP-ARNQLVAAMLAVENDASLTQHDKARKRQELLTGKAPADDDDDDEQENKSSL 376
Query: 117 LDIFDGSINCSFCMQLPDR 135
DI S+NCS C+QLP+R
Sbjct: 377 SDILSRSLNCSICIQLPER 433
>TC222633
Length = 709
Score = 79.3 bits (194), Expect = 5e-15
Identities = 38/75 (50%), Positives = 61/75 (80%)
Frame = +1
Query: 519 EGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQ 578
EG F+G++F+++R+ +GGR+L +QKNVMKCPSCS DI+D+LQN QV+ ++ +ESL+++
Sbjct: 7 EGEFAGQAFVKERS-KGGRTLLSQKNVMKCPSCSIDISDYLQNIQVDIDLKSAIESLKAK 183
Query: 579 IAQDENSEQVENSEE 593
+ +E E VE+SE+
Sbjct: 184 V--EEIGESVESSED 222
>TC228188 similar to GB|AAP31929.1|30387527|BT006585 At3g58030 {Arabidopsis
thaliana;} , partial (27%)
Length = 877
Score = 45.8 bits (107), Expect = 6e-05
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 2/94 (2%)
Frame = +1
Query: 80 RAIDSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILDIFDGSINCSFCMQLPDRPVTT 139
+A D+ + + + ++KK+++ G +G D FD C+ C+ L PV T
Sbjct: 142 KACDNINGVAEDETSQKKEDIEKG---------SGNDGDFFD----CNICLDLARDPVVT 282
Query: 140 PCGHNFCLKCFEKW--IGQGKRTCANCRRDIPAK 171
CGH FC C +W + + C C+ ++ K
Sbjct: 283 CCGHLFCWPCLYRWLHLHSDAKECPVCKGEVTLK 384
Score = 38.5 bits (88), Expect = 0.010
Identities = 29/139 (20%), Positives = 53/139 (37%), Gaps = 8/139 (5%)
Frame = +1
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSC 551
F CNIC + P+ T C H FC CL R L + +CP C
Sbjct: 235 FDCNICLDLARDPVVTCCGHLFCWPCLY-----------------RWLHLHSDAKECPVC 363
Query: 552 SNDIADFLQNPQVNR--EMMGVVESLQSQIAQDENSEQVENSEELSDKS------DEIVK 603
++ P R + G E +I +++VE+ + ++ +E+++
Sbjct: 364 KGEVTLKSVTPIYGRANNVRGPEEDSALKIPPRPQAKRVESLRQTIQRNAFALPVEEMIR 543
Query: 604 NVPDEVEVSKPCDDSTESD 622
+ +E+++ E D
Sbjct: 544 RLGSRIELTRDLVQPPEPD 600
>TC216628 weakly similar to UP|Q8H0X2 (Q8H0X2) Expressed protein (At1g18660),
partial (50%)
Length = 872
Score = 44.3 bits (103), Expect = 2e-04
Identities = 15/28 (53%), Positives = 21/28 (74%)
Frame = +2
Query: 491 EFGCNICHKVLSSPLTTPCAHNFCKVCL 518
+F C +C K+L P+TTPC H+FC+ CL
Sbjct: 575 DFDCTLCLKLLYEPVTTPCGHSFCRSCL 658
Score = 42.0 bits (97), Expect = 9e-04
Identities = 15/33 (45%), Positives = 23/33 (69%)
Frame = +2
Query: 125 NCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQG 157
+C+ C++L PVTTPCGH+FC C + + +G
Sbjct: 581 DCTLCLKLLYEPVTTPCGHSFCRSCLFQSMDRG 679
>CO982502
Length = 677
Score = 44.3 bits (103), Expect = 2e-04
Identities = 18/47 (38%), Positives = 22/47 (46%)
Frame = -1
Query: 122 GSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCRRDI 168
G C+ C+ P P T CGH FC C +W + K C CR I
Sbjct: 248 GVSKCTLCLSNPQHPTATSCGHVFCWNCITEWCNE-KPECPLCRTPI 111
>BE802170
Length = 407
Score = 43.9 bits (102), Expect = 2e-04
Identities = 27/94 (28%), Positives = 42/94 (43%), Gaps = 8/94 (8%)
Frame = +2
Query: 83 DSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILDIFDGSINCSFCMQLPDRPVTTPCG 142
DS+ ++ +QK+K S +P + D FD C+ CM+ PV T CG
Sbjct: 158 DSNGEVSLKQKSK---------SISAEPTISTSEYDCFD----CNICMESAHDPVVTLCG 298
Query: 143 HNFCLKCFEKWIG--------QGKRTCANCRRDI 168
H +C C KW+ ++TC C+ +I
Sbjct: 299 HLYCWPCIYKWLDVQSSSVEPYQQQTCPVCKSEI 400
Score = 32.3 bits (72), Expect = 0.71
Identities = 14/46 (30%), Positives = 24/46 (51%), Gaps = 4/46 (8%)
Frame = +2
Query: 477 KKTKNVSLKEKLLKE----FGCNICHKVLSSPLTTPCAHNFCKVCL 518
+K+K++S + + F CNIC + P+ T C H +C C+
Sbjct: 185 QKSKSISAEPTISTSEYDCFDCNICMESAHDPVVTLCGHLYCWPCI 322
>TC216036 similar to GB|AAP13393.1|30023720|BT006285 At4g27470 {Arabidopsis
thaliana;} , partial (23%)
Length = 1061
Score = 43.5 bits (101), Expect = 3e-04
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = +3
Query: 111 PAAGGILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWI 154
P+ + + DG +C+ C+ PV T CGH +C C KW+
Sbjct: 237 PSPMTVTENLDGCFDCNICLDFAHEPVVTLCGHLYCWPCIYKWL 368
Score = 32.3 bits (72), Expect = 0.71
Identities = 22/85 (25%), Positives = 32/85 (36%)
Frame = +3
Query: 482 VSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRA 541
+++ E L F CNIC P+ T C H +C C+ +S + A
Sbjct: 246 MTVTENLDGCFDCNICLDFAHEPVVTLCGHLYCWPCIYKWLHVQS-----------ASLA 392
Query: 542 QKNVMKCPSCSNDIADFLQNPQVNR 566
+CP C +DI P R
Sbjct: 393 PDEHPQCPVCKDDICHTTMVPLYGR 467
>TC227743 similar to PIR|E86326|E86326 protein F18O14.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(73%)
Length = 1212
Score = 43.1 bits (100), Expect = 4e-04
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 5/68 (7%)
Frame = +2
Query: 103 GPSSGEKPPAAGGILDIFDG---SINCSFCMQLPDRPVTTPCGHNFCLKCFEKWI--GQG 157
G S+G PP+ + + + C+ C +L P+ T CGH FC C KW+
Sbjct: 140 GESTGRSPPSPSYFNNNNNNDAANFECNICFELAQDPIITLCGHLFCWPCLYKWLHFHSQ 319
Query: 158 KRTCANCR 165
R C C+
Sbjct: 320 SRECPVCK 343
Score = 35.0 bits (79), Expect = 0.11
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +2
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCL 518
F CNIC ++ P+ T C H FC CL
Sbjct: 212 FECNICFELAQDPIITLCGHLFCWPCL 292
>TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing protein,
partial (51%)
Length = 1103
Score = 42.4 bits (98), Expect = 7e-04
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 9/56 (16%)
Frame = +1
Query: 122 GSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIG---------QGKRTCANCRRDI 168
G +C+ C++ PV T CGH +C C KW+ + K+ C C+ +I
Sbjct: 235 GGFDCNICLECVQDPVVTLCGHLYCWPCIYKWLNLQTASSENEEEKQQCPVCKSEI 402
Score = 33.9 bits (76), Expect = 0.24
Identities = 16/65 (24%), Positives = 30/65 (45%)
Frame = +1
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSC 551
F CNIC + + P+ T C H +C C+ + ++ S ++ +CP C
Sbjct: 241 FDCNICLECVQDPVVTLCGHLYCWPCIYKWLNLQT----------ASSENEEEKQQCPVC 390
Query: 552 SNDIA 556
++I+
Sbjct: 391 KSEIS 405
>AW755410
Length = 433
Score = 41.6 bits (96), Expect = 0.001
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = +1
Query: 122 GSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWI 154
GS +C+ C++ PV T CGH +C C KW+
Sbjct: 283 GSFDCNICLECVQDPVVTLCGHLYCWPCIYKWL 381
Score = 32.7 bits (73), Expect = 0.54
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +1
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCL 518
F CNIC + + P+ T C H +C C+
Sbjct: 289 FDCNICLECVQDPVVTLCGHLYCWPCI 369
>TC216035 similar to GB|AAP13393.1|30023720|BT006285 At4g27470 {Arabidopsis
thaliana;} , partial (32%)
Length = 966
Score = 41.2 bits (95), Expect = 0.002
Identities = 24/87 (27%), Positives = 34/87 (38%), Gaps = 8/87 (9%)
Frame = +1
Query: 121 DGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRT--------CANCRRDIPAKI 172
+G +C+ C+ PV T CGH +C C KW+ + C C+ DI
Sbjct: 190 NGCFDCNICLDFAHEPVVTLCGHLYCWPCIYKWLHVQSDSLAPDEHPQCPVCKADIS--- 360
Query: 173 ASQPRINSQLAIAIRLAKLAKSEGKVS 199
NS + A +EGK S
Sbjct: 361 ------NSTMVPLYGRGHAATAEGKTS 423
Score = 32.0 bits (71), Expect = 0.92
Identities = 22/86 (25%), Positives = 29/86 (33%)
Frame = +1
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSC 551
F CNIC P+ T C H +C C+ +S A +CP C
Sbjct: 199 FDCNICLDFAHEPVVTLCGHLYCWPCIYKWLHVQS-----------DSLAPDEHPQCPVC 345
Query: 552 SNDIADFLQNPQVNREMMGVVESLQS 577
DI++ P R E S
Sbjct: 346 KADISNSTMVPLYGRGHAATAEGKTS 423
>TC217261
Length = 1186
Score = 41.2 bits (95), Expect = 0.002
Identities = 35/106 (33%), Positives = 43/106 (40%), Gaps = 19/106 (17%)
Frame = +3
Query: 85 DASLTDQQKAKKKQE--------------LVGGPSSGE----KPPAAGGILDIFDGSINC 126
D L DQ + K++ L GG SS E KPP L NC
Sbjct: 366 DVDLEDQTRRNKRRRELPNLIINCDKYINLEGGSSSMEQSFKKPPEPPKEL-----VFNC 530
Query: 127 SFCMQLPDRPVTTPCGHNFCLKCFEKWI-GQGKRTCANCRRDIPAK 171
CM ++T CGH FC C + I QGK C CR+ + AK
Sbjct: 531 PICMGPMVHEMSTRCGHIFCKDCIKAAISAQGK--CPTCRKKVVAK 662
Score = 34.3 bits (77), Expect = 0.19
Identities = 17/64 (26%), Positives = 25/64 (38%)
Frame = +3
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSC 551
F C IC + ++T C H FCK C++ A S + KCP+C
Sbjct: 522 FNCPICMGPMVHEMSTRCGHIFCKDCIKAAISAQG--------------------KCPTC 641
Query: 552 SNDI 555
+
Sbjct: 642 RKKV 653
>TC218937 similar to UP|PEXA_ARATH (Q9SYU4) Peroxisome assembly protein 10
(Peroxin-10) (AthPEX10) (Pex10p) (PER8), partial (73%)
Length = 1217
Score = 40.8 bits (94), Expect = 0.002
Identities = 17/47 (36%), Positives = 21/47 (44%)
Frame = +1
Query: 122 GSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCRRDI 168
G C+ C+ P T CGH FC C +W + K C CR I
Sbjct: 946 GVSKCTLCLSNRQHPTATSCGHVFCWNCITEWCNE-KPECPLCRTPI 1083
>TC206224 similar to PIR|E86326|E86326 protein F18O14.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(43%)
Length = 1080
Score = 40.4 bits (93), Expect = 0.003
Identities = 16/44 (36%), Positives = 20/44 (45%)
Frame = +1
Query: 111 PAAGGILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWI 154
P+ G G C+ C L PV T CGH FC C +W+
Sbjct: 271 PSCSGNSSNDAGDFECNICFDLAQDPVITLCGHLFCWPCLYRWL 402
Score = 34.7 bits (78), Expect = 0.14
Identities = 13/28 (46%), Positives = 16/28 (56%)
Frame = +1
Query: 491 EFGCNICHKVLSSPLTTPCAHNFCKVCL 518
+F CNIC + P+ T C H FC CL
Sbjct: 307 DFECNICFDLAQDPVITLCGHLFCWPCL 390
>TC227914 similar to UP|Q6R567 (Q6R567) Ring domain containing protein,
partial (29%)
Length = 1078
Score = 40.0 bits (92), Expect = 0.003
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +1
Query: 122 GSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWI 154
G +C+ C++ PV T CGH +C C KW+
Sbjct: 283 GDFDCNICLECVQDPVVTLCGHLYCWPCIYKWL 381
Score = 33.5 bits (75), Expect = 0.32
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +1
Query: 491 EFGCNICHKVLSSPLTTPCAHNFCKVCL 518
+F CNIC + + P+ T C H +C C+
Sbjct: 286 DFDCNICLECVQDPVVTLCGHLYCWPCI 369
>TC221337 similar to UP|Q9SD57 (Q9SD57) RNA-binding protein-like protein,
partial (77%)
Length = 1151
Score = 40.0 bits (92), Expect = 0.003
Identities = 13/42 (30%), Positives = 24/42 (56%)
Frame = +1
Query: 124 INCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCR 165
+ C C+++ + V C H+ C+KC+E W + ++C CR
Sbjct: 748 LECGICLEINSKVVLPNCNHSMCMKCYEDWHAR-SQSCPFCR 870
>TC216627 similar to UP|Q8H0X2 (Q8H0X2) Expressed protein (At1g18660),
partial (51%)
Length = 1037
Score = 39.7 bits (91), Expect = 0.004
Identities = 16/31 (51%), Positives = 21/31 (67%)
Frame = +1
Query: 135 RPVTTPCGHNFCLKCFEKWIGQGKRTCANCR 165
RPVTTPCGH+FC C + + +G + C CR
Sbjct: 7 RPVTTPCGHSFCCSCLFQSMDRGNK-CPLCR 96
Score = 30.0 bits (66), Expect = 3.5
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +1
Query: 504 PLTTPCAHNFCKVCL 518
P+TTPC H+FC CL
Sbjct: 10 PVTTPCGHSFCCSCL 54
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,659,578
Number of Sequences: 63676
Number of extensions: 651243
Number of successful extensions: 3815
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 3666
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3802
length of query: 680
length of database: 12,639,632
effective HSP length: 104
effective length of query: 576
effective length of database: 6,017,328
effective search space: 3465980928
effective search space used: 3465980928
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0191.7


