

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
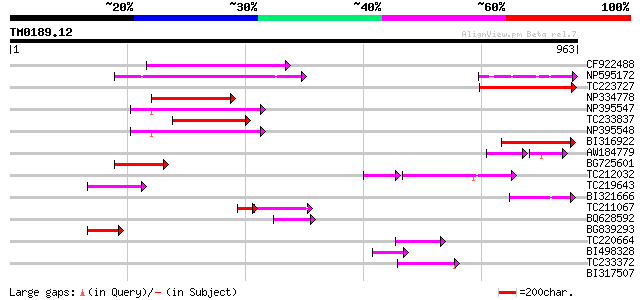
Query= TM0189.12
(963 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922488 198 1e-50
NP595172 polyprotein [Glycine max] 179 4e-45
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 162 9e-40
NP334778 reverse transcriptase [Glycine max] 148 1e-35
NP395547 reverse transcriptase [Glycine max] 145 6e-35
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 142 7e-34
NP395548 reverse transcriptase [Glycine max] 133 3e-31
BI316922 124 3e-28
AW184779 67 2e-21
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 100 2e-21
TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 79 6e-20
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 81 3e-15
BI321666 78 2e-14
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 47 3e-08
BQ628592 55 2e-07
BG839293 52 1e-06
TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial... 49 8e-06
BI498328 46 9e-05
TC233372 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial... 45 2e-04
BI317507 42 0.002
>CF922488
Length = 741
Score = 198 bits (503), Expect = 1e-50
Identities = 107/245 (43%), Positives = 145/245 (58%)
Frame = +3
Query: 233 VKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQ 292
V K +GK MCVD DLN PKD +PLP I+ LVD S MD +SGY+QIK+
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 293 SDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVL 352
D +KTTF+T +CY+ M FGLKN GATYQR M +F + + +EVY+DDMIVKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 353 GSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMK 412
H +L + F R++K+ +RLNP KC F ++ K L F+ + R IE++ +K K I EM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 413 SPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLS 472
P K+VQ +GR+ + RF+ P L KN +W+ +C AF R+K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 473 APPVL 477
P VL
Sbjct: 723 NPHVL 737
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 179 bits (455), Expect = 4e-45
Identities = 99/325 (30%), Positives = 174/325 (53%)
Frame = +1
Query: 179 HRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG 238
H + L G P+ R + ++ I++ + ++L I+ P + + +++VKK +G
Sbjct: 1759 HAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSP-FSLPILLVKKKDG 1935
Query: 239 KWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKT 298
WR C D LN KDS+P+P++D+L+D G + S +D SGYHQI + D +KT
Sbjct: 1936 SWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKT 2115
Query: 299 TFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHE 358
F T +Y + MPFGL NA AT+Q LM+++F+ + + + V+ DD+++ S +H +
Sbjct: 2116 AFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLK 2295
Query: 359 DLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVK 418
L +++H + KCSFG +LG ++ + + K +A+ + +P+NVK
Sbjct: 2296 HLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVK 2475
Query: 419 EVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLS 478
+++ +G RF+ + + P L+K+ +F WN+E E AF +LK+ ++ PVLS
Sbjct: 2476 QLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKD-SFLWNNEAEAAFVKLKKAMTEAPVLS 2652
Query: 479 KPVQGLPLHLYFSVGDHAISSVILQ 503
P P L + +V+ Q
Sbjct: 2653 LPDFSQPFILETDASGIGVGAVLGQ 2727
Score = 62.8 bits (151), Expect = 7e-10
Identities = 49/175 (28%), Positives = 84/175 (48%), Gaps = 8/175 (4%)
Frame = +1
Query: 797 QRREAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAV---MTEVHEGVCASHIG-GRSLAS 852
Q +A HYT+ +G+L+ + R + P + E V + E H H G R+LA
Sbjct: 3124 QGADASHYTVREGLLYWKD------RVVIPAEEEIVNKILQEYHSSPIGGHAGITRTLAR 3285
Query: 853 KVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMW---GVDLV 909
L+A FYWP ++++ +++KC CQ P G L + P P +W +D +
Sbjct: 3286 --LKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPL--PIPQQVWEDVAMDFI 3453
Query: 910 GPFPTARSQMKFILVAVDYFTKWVEAEPLASITAAKIIS-FYWKRIVCRFGVPRA 963
P + + I+V +D TK+ PL + +K+++ + IV G+PR+
Sbjct: 3454 TGLPNSFG-LSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRS 3615
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 162 bits (409), Expect = 9e-40
Identities = 74/166 (44%), Positives = 103/166 (61%), Gaps = 1/166 (0%)
Frame = +1
Query: 798 RREAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRA 857
RR A + + IL++R LRC+ + ++ EVHEG +H G ++A K+LRA
Sbjct: 217 RRLAAGFFMSGSILYKRNHDMKPLRCVDAREANHMIEEVHEGSFGTHANGHAMARKILRA 396
Query: 858 GFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMWGVDLVGPF-PTAR 916
G+YW T+ +C VRKC KCQ F D APP L +SSPWPF+MWG+D++G P A
Sbjct: 397 GYYWLTMESDCCVHVRKCHKCQAFADNVNAPPHPLNVMSSPWPFSMWGIDVIGAIEPKAS 576
Query: 917 SQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVPR 962
+ +FILVA+DYFTKWVEA + ++ F K I+CR+G+PR
Sbjct: 577 NGHRFILVAIDYFTKWVEAASYTDVMRGVVVRFIKKEIICRYGLPR 714
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 148 bits (373), Expect = 1e-35
Identities = 70/143 (48%), Positives = 94/143 (64%)
Frame = +3
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
RMCVD DLN+ PKD++PLP ID L+ L S MD +SGY+QIKM D +KTTF
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
+T +CY+ M FGLKN GATY R M +F+ + + +E YVD+MI KS + H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 361 MEAFGRIQKHNMRLNPEKCSFGI 383
FG+++K+ +RLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 145 bits (367), Expect = 6e-35
Identities = 83/248 (33%), Positives = 125/248 (49%), Gaps = 18/248 (7%)
Frame = +1
Query: 205 IQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG------------------KWRMCVDN 246
+++EV KLL A I I +W+ V +V K G +WRMC+D
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 247 TDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMTARVN 306
LN+ KD YPLP +D+++ +D YSGY+QI + D++KT F
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 307 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFGR 366
+ Y+ MPFGL NA T+QR M +F+ V + +EV++DD N +L + R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 367 IQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGR 426
+K N+ LN EKC F ++ G LG I+ R IE+ +K I ++ P NVK + +G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 427 ITALSRFL 434
+ RF+
Sbjct: 721 VGFYRRFI 744
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 142 bits (358), Expect = 7e-34
Identities = 66/132 (50%), Positives = 94/132 (71%)
Frame = +2
Query: 277 SLMDAYSGYHQIKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVG 336
S MD +SGY+QI M + D +KTTF+T + Y+ M FGLKN GATYQR M +F +
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMMH 184
Query: 337 RNMEVYVDDMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSR 396
+ +EVYVDDMI KS + H +L + FGR+QK+ ++LNP KC+FG++ GK LGF+++ +
Sbjct: 185 KEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQK 364
Query: 397 RIEINPDKCKAI 408
IEI+P+K KA+
Sbjct: 365 GIEIDPEKVKAL 400
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 133 bits (335), Expect = 3e-31
Identities = 78/248 (31%), Positives = 127/248 (50%), Gaps = 18/248 (7%)
Frame = +1
Query: 205 IQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG------------------KWRMCVDN 246
+++EV KLL I I W+ V++V K G W++C+D
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 247 TDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMTARVN 306
LN+ KD +PLP +D++++ G+ +DAY GY+QI + D++K F
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 307 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFGR 366
+ Y+ +PFGL NA T+Q M +F V +++EV++DD V + + L R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 367 IQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGR 426
+ N+ LN EKC F +R G LG I++R IE++ K I+++ PSNVK ++ +G+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 427 ITALSRFL 434
RF+
Sbjct: 721 ARFYRRFI 744
>BI316922
Length = 405
Score = 124 bits (310), Expect = 3e-28
Identities = 54/127 (42%), Positives = 85/127 (66%)
Frame = +3
Query: 835 EVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVT 894
E+H G+C H + + ++VLR G+Y T+RK C E+V+KC++C F ++ +L
Sbjct: 3 EMHRGICGMHSKSQLMTTRVLRVGYY**TMRKYCTEYVKKCEEC*KFGNISHLLVEELHN 182
Query: 895 ISSPWPFAMWGVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRI 954
I +PWPFA+ GVD++ PFP ++ Q+K++LV +D FTKW+E E +A I+ A + F + I
Sbjct: 183 IVAPWPFAI*GVDILRPFPLSKRQVKYLLVGIDQFTKWIETEHIAIISIANVRKFV*RNI 362
Query: 955 VCRFGVP 961
VC FG+P
Sbjct: 363 VC*FGIP 383
>AW184779
Length = 432
Score = 67.4 bits (163), Expect(2) = 2e-21
Identities = 29/70 (41%), Positives = 42/70 (59%)
Frame = +1
Query: 810 ILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECA 869
IL++R LLRC+ + E ++ EVHEG +H ++A K+LR G+YW T+ +C
Sbjct: 13 ILYKRNHDMVLLRCVDAREAEQMLVEVHEGSFGTHANIHAMAQKILRVGYYWLTMESDCC 192
Query: 870 EFVRKCKKCQ 879
V KC KCQ
Sbjct: 193 IHVWKCHKCQ 222
Score = 54.3 bits (129), Expect(2) = 2e-21
Identities = 29/75 (38%), Positives = 43/75 (56%), Gaps = 10/75 (13%)
Frame = +3
Query: 883 DLPRAPPGQLVTISSPWPFA---------MWGVDLVGPF-PTARSQMKFILVAVDYFTKW 932
++P P + + +P P+ MWG+D++G P A + FILVA+DYFTKW
Sbjct: 204 EMP*VPVRPSLIMLTPHPYL*MSWQHLGHMWGIDVIGAIEPKASNGHHFILVAIDYFTKW 383
Query: 933 VEAEPLASITAAKII 947
VEA AS+T + +I
Sbjct: 384 VEAVSYASVTRSVVI 428
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 100 bits (250), Expect = 2e-21
Identities = 48/91 (52%), Positives = 65/91 (70%)
Frame = -3
Query: 179 HRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG 238
H+LA+ V+ +TQ +R++ E+ + + QEV KL A FIR+I Y T L +VV+VKK NG
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 239 KWRMCVDNTDLNKTCPKDSYPLPSIDKLVDG 269
KWR+C D DLN CPKD+YPLP+ID + DG
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTDG 5
>TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (3%)
Length = 803
Score = 79.3 bits (194), Expect(2) = 6e-20
Identities = 60/201 (29%), Positives = 95/201 (46%), Gaps = 7/201 (3%)
Frame = +2
Query: 668 LKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPR 727
++ AI+ + L V DS LV +Q+ GE + ++ LI Y ++ L G ++ V
Sbjct: 206 VQAAIDSNVKLLKVYGDSALVIHQLRGECETRDPNLIPYQAYIKELAGFFDEISFHHVA* 385
Query: 728 AQNQRADALAKLASTRK--PGNNRSVIQETLANPNIEGEFVASVDRQETWMNPIIDILAG 785
+NQ ADALA L S + P + I+ V + W I +
Sbjct: 386 EENQMADALATLVSMFQLTPHGDLPYIEFRCRGRPAHCCLVEEERDGKPWYFDIKRYVES 565
Query: 786 -----EPSDVVKYSKAQRREAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGV 840
E SD K + +RR A + + IL++R LL C+ ++ E ++ EVHEG
Sbjct: 566 KEYPLEASDNDK--RRKRRLAAGFFMSGSILYKRNHDMVLLHCVNGKEVENMLGEVHEGS 739
Query: 841 CASHIGGRSLASKVLRAGFYW 861
+H G ++A K+LRAG+YW
Sbjct: 740 FGTHSNGHAMARKILRAGYYW 802
Score = 37.4 bits (85), Expect(2) = 6e-20
Identities = 20/62 (32%), Positives = 32/62 (51%)
Frame = +1
Query: 602 LVELTPEEGEKVSTQWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTSNNQAEY 661
++ L E+ ++ +WI++ D +SN G G L P + + R F +NN AEY
Sbjct: 7 IMALFEEKLDEDRDKWIVWFDRASNVLGHGVGAILVSPDNQCIPFTTRLGFDCTNNMAEY 186
Query: 662 EA 663
EA
Sbjct: 187 EA 192
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 80.9 bits (198), Expect = 3e-15
Identities = 39/99 (39%), Positives = 59/99 (59%)
Frame = +3
Query: 133 GEKILKIGTKLSEEQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLALNPGVEPITQ 192
G++ +KIGT ++ + + LL D D+FAWS++ MPG+ + + HRL LNP P+ Q
Sbjct: 1014 GKREVKIGTGITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNPECSPVKQ 1193
Query: 193 TRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVV 231
RRM E I++EV K A F+ +YP W+ N+V
Sbjct: 1194 KLRRMKPETSLKIKEEVKK*FDAGFLAVARYPKWVANIV 1310
>BI321666
Length = 430
Score = 78.2 bits (191), Expect = 2e-14
Identities = 44/112 (39%), Positives = 61/112 (54%)
Frame = +2
Query: 850 LASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMWGVDLV 909
+++ VL++ FY P+I K+ + C KCQ + + L TI F WG+D V
Sbjct: 2 ISTNVLQSRFYLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFV 181
Query: 910 GPFPTARSQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVP 961
GPFP + S ++ILV VDY +KWVEA A +I F K+I R GVP
Sbjct: 182 GPFPPSFSN-EYILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVP 334
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 46.6 bits (109), Expect(2) = 3e-08
Identities = 29/99 (29%), Positives = 47/99 (47%)
Frame = +1
Query: 416 NVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPP 475
+V +++ G + RF+ + ++P + +KKN F W + E+AF LKE L+ P
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 476 VLSKPVQGLPLHLYFSVGDHAISSVILQEVDGEQKIVYF 514
VL+ P L + +V+LQ G I YF
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQ---GGHPIAYF 396
Score = 30.4 bits (67), Expect(2) = 3e-08
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +2
Query: 388 FLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
F GF++ ++++P+K KAIQE P V E+
Sbjct: 23 FSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>BQ628592
Length = 423
Score = 54.7 bits (130), Expect = 2e-07
Identities = 27/74 (36%), Positives = 44/74 (58%), Gaps = 2/74 (2%)
Frame = -1
Query: 448 LKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQEVDG 507
L KN WNS +EAF ++K+ L+ P VL PV G P LY ++ D ++ V++Q D
Sbjct: 423 LPKNQAILWNSNYQEAFEKIKQSLANPSVLMPPVTGRPFLLYMTMLDESMGCVLVQHDDS 244
Query: 508 --EQKIVYFVSYHF 519
+++ +Y++S F
Sbjct: 243 GKKEQAIYYLSKKF 202
>BG839293
Length = 781
Score = 52.0 bits (123), Expect = 1e-06
Identities = 23/60 (38%), Positives = 38/60 (63%)
Frame = +1
Query: 133 GEKILKIGTKLSEEQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLALNPGVEPITQ 192
G++ +KIGT ++ + + LL D D+FAWS++ MPG+ + + H+L LNP P+ Q
Sbjct: 550 GKREVKIGTGITAPIREELIILLKDYQDIFAWSYQDMPGLSSDIVQHQLPLNPECSPVKQ 729
>TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (31%)
Length = 724
Score = 49.3 bits (116), Expect = 8e-06
Identities = 29/86 (33%), Positives = 44/86 (50%)
Frame = +1
Query: 655 SNNQAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLM 714
+NN AEY A+I G+K A++ + ++ DS LV Q++G ++VK L + L
Sbjct: 64 TNNAAEYRAMILGMKYALKKGFTGIRIQGDSKLVCMQIDGSWKVKNENLSTLYNVAKELK 243
Query: 715 GRLQQVVVEFVPRAQNQRADALAKLA 740
+ + V R N ADA A LA
Sbjct: 244 DKFSSFQISHVLRNFNSDADAQANLA 321
>BI498328
Length = 335
Score = 45.8 bits (107), Expect = 9e-05
Identities = 22/61 (36%), Positives = 33/61 (54%)
Frame = +1
Query: 616 QWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTSNNQAEYEALIAGLKLAIEVQ 675
+WI+ DG+SN G G L P + + + R F +NN AEYEA G++ AI+
Sbjct: 148 KWIVCFDGASNALGHGVGAVLVSPDDQCIPFTARLGFDCTNNMAEYEACALGVQAAIDFD 327
Query: 676 I 676
+
Sbjct: 328 V 330
>TC233372 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (12%)
Length = 561
Score = 44.7 bits (104), Expect = 2e-04
Identities = 33/109 (30%), Positives = 48/109 (43%), Gaps = 4/109 (3%)
Frame = +1
Query: 659 AEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQ 718
AEY +LI GL + + V+ DS LV NQ+ G +++K + + L +
Sbjct: 1 AEYRSLILGLXHXXKKGYKXIIVQGDSLLVCNQIQGLWKIKNQNMGTLCXEAKELKDKFL 180
Query: 719 QVVVEFVPRAQNQRADALAKLASTRKPGNNRSVIQ----ETLANPNIEG 763
+ +PR N ADA A LA + + Q TLA PN G
Sbjct: 181 SFKISHIPREYNSEADAQANLAINLRACEVQEDCQLL*CSTLAVPNYLG 327
>BI317507
Length = 359
Score = 41.6 bits (96), Expect = 0.002
Identities = 30/113 (26%), Positives = 54/113 (47%), Gaps = 1/113 (0%)
Frame = -1
Query: 345 DMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDK 404
++++ S +H L ++K + N +KC F ++LG +I+ + ++ +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 405 CKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPF-IKCLKKNTTFEW 456
K++ E P NVK V + R+T R + K AP + L KN F+W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFL-RLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKW 18
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,074,610
Number of Sequences: 63676
Number of extensions: 576751
Number of successful extensions: 2478
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 2454
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2470
length of query: 963
length of database: 12,639,632
effective HSP length: 106
effective length of query: 857
effective length of database: 5,889,976
effective search space: 5047709432
effective search space used: 5047709432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0189.12


