

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
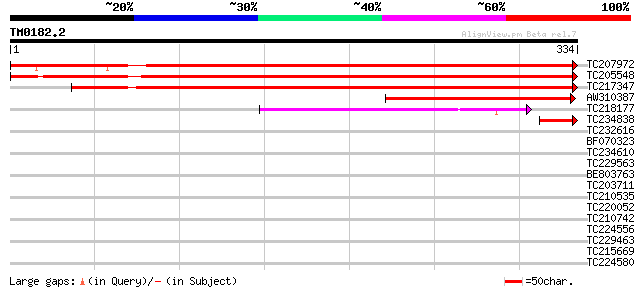
Query= TM0182.2
(334 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207972 UP|Q7XZQ0 (Q7XZQ0) Alternative oxidase 2a, complete 531 e-151
TC205548 UP|Q7XZQ1 (Q7XZQ1) Alternative oxidase 2b, complete 447 e-126
TC217347 homologue to UP|AOX1_SOYBN (Q07185) Alternative oxidase... 433 e-122
AW310387 similar to SP|O03376|AOX3 Alternative oxidase 3 mitoch... 157 5e-39
TC218177 similar to PIR|T52422|T52422 alternative oxidase-relate... 76 2e-14
TC234838 UP|AOX2_SOYBN (Q41266) Alternative oxidase 2, mitochond... 45 6e-05
TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|0... 32 0.53
BF070323 32 0.53
TC234610 UP|Q9M6B9 (Q9M6B9) Isoflavone synthase 1 (Fragment), pa... 30 1.5
TC229563 30 2.0
BE803763 weakly similar to SP|Q41266|AOX2 Alternative oxidase 2 ... 29 2.6
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 29 3.4
TC210535 similar to GB|AAR28019.1|39545912|AY463617 TAF14b {Arab... 29 3.4
TC220052 similar to GB|AAM51598.1|21436041|AY116964 AT3g47850/T2... 28 4.4
TC210742 similar to GB|AAM78055.1|21928053|AY125545 At4g29227 {A... 28 4.4
TC224556 weakly similar to UP|Q9LWR5 (Q9LWR5) ESTs AU069293(C539... 28 7.6
TC229463 weakly similar to UP|O81329 (O81329) F3D13.5 protein, p... 28 7.6
TC215669 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, part... 28 7.6
TC224580 similar to UP|Q6Y1D6 (Q6Y1D6) 60S acidic ribosomal prot... 28 7.6
>TC207972 UP|Q7XZQ0 (Q7XZQ0) Alternative oxidase 2a, complete
Length = 1366
Score = 531 bits (1367), Expect = e-151
Identities = 268/343 (78%), Positives = 286/343 (83%), Gaps = 9/343 (2%)
Frame = +3
Query: 1 MKHLALSYALRRAL------NCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQ 54
MK AL+ +RRAL N NR G A+ A E R G NG F W R M S
Sbjct: 81 MKLTALNSTVRRALLNGRNQNGNRLGSAALMPYAAAETRLLCAGGANGWFFYWKRTMVSP 260
Query: 55 AA---PEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPW 111
A PE+EK++EKA+ E SVV SSYWGISRPK+ REDGTEWPWNCFMPW
Sbjct: 261 AEAKLPEKEKEKEKAKAEK----------SVVESSYWGISRPKVVREDGTEWPWNCFMPW 410
Query: 112 ETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGM 171
E+YR ++SIDLTKHHVPKN LDKVAYRTVKLLRIPTD+FF+RRYGCRAMMLETVAAVPGM
Sbjct: 411 ESYRSNVSIDLTKHHVPKNVLDKVAYRTVKLLRIPTDLFFKRRYGCRAMMLETVAAVPGM 590
Query: 172 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFF 231
VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELV+PKWYER LVL VQGVFF
Sbjct: 591 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVKPKWYERLLVLAVQGVFF 770
Query: 232 NAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA 291
NAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKD+ESGAIENVPAPAIAIDYWRLPKDA
Sbjct: 771 NAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDLESGAIENVPAPAIAIDYWRLPKDA 950
Query: 292 TLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 951 RLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 1079
>TC205548 UP|Q7XZQ1 (Q7XZQ1) Alternative oxidase 2b, complete
Length = 1384
Score = 447 bits (1149), Expect = e-126
Identities = 218/335 (65%), Positives = 259/335 (77%), Gaps = 1/335 (0%)
Frame = +2
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWN-RMMSSQAAPEE 59
MK++ + A R L G + RQL + G F ++ R MS+ ++
Sbjct: 137 MKNVLVRSAARALLGGG--GRSYYRQLSTAAIVEQRHQHGGGAFGSFHLRRMSTLPEVKD 310
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
+ EEK + + + + VV+SYWGI+RPK+ REDGTEWPWNCFMPW++Y D+S
Sbjct: 311 QHSEEKKNEVN-------DTSNAVVTSYWGITRPKVRREDGTEWPWNCFMPWDSYHSDVS 469
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
ID+TKHH PK+ DKVA+R VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL
Sbjct: 470 IDVTKHHTPKSLTDKVAFRAVKFLRVLSDIYFKERYGCHAMMLETIAAVPGMVGGMLLHL 649
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
+SLRKFQ SGGWIKALLEEAENERMHLMTMVELV+P W+ER L+ T QGVFFNAFFV YL
Sbjct: 650 KSLRKFQHSGGWIKALLEEAENERMHLMTMVELVKPSWHERLLIFTAQGVFFNAFFVFYL 829
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
LSPK AHR VGYLEEEA+ SYT++L IESG +ENVPAPAIAIDYWRLPKDATLKDV+TV
Sbjct: 830 LSPKAAHRFVGYLEEEAVISYTQHLNAIESGKVENVPAPAIAIDYWRLPKDATLKDVVTV 1009
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
IRADEAHHRDVNHFASDIH GKEL+EAPAP+GYH
Sbjct: 1010IRADEAHHRDVNHFASDIHHQGKELKEAPAPIGYH 1114
>TC217347 homologue to UP|AOX1_SOYBN (Q07185) Alternative oxidase 1,
mitochondrial precursor , complete
Length = 1709
Score = 433 bits (1113), Expect = e-122
Identities = 208/298 (69%), Positives = 242/298 (80%)
Frame = +1
Query: 37 VSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKIT 96
V G ++ R S+ A E+EK E+K S A N V+ SYWGI KIT
Sbjct: 205 VGGLRALYGGGVRSESTLALSEKEKIEKKVGLSS----AGGNKEEKVIVSYWGIQPSKIT 372
Query: 97 REDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYG 156
++DGTEW WNCF PWETY+ DLSIDL KHH P FLDK+A+ TVK+LR PTDVFFQRRYG
Sbjct: 373 KKDGTEWKWNCFRPWETYKADLSIDLEKHHAPTTFLDKMAFWTVKVLRYPTDVFFQRRYG 552
Query: 157 CRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPK 216
CRAMMLETVAAVPGMV GMLLH +SLR+F+ SGGWIKALLEEAENERMHLMT +E+ +PK
Sbjct: 553 CRAMMLETVAAVPGMVAGMLLHCKSLRRFEHSGGWIKALLEEAENERMHLMTFMEVAKPK 732
Query: 217 WYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVP 276
WYER LV+TVQGVFFNA+F+ YLLSPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVP
Sbjct: 733 WYERALVITVQGVFFNAYFLGYLLSPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVP 912
Query: 277 APAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
APAIAIDYW+LP +TL+DV+ V+RADEAHHRDVNHFASDIH+ G+ELREA AP+GYH
Sbjct: 913 APAIAIDYWQLPPGSTLRDVVMVVRADEAHHRDVNHFASDIHYQGRELREAAAPIGYH 1086
>AW310387 similar to SP|O03376|AOX3 Alternative oxidase 3 mitochondrial
precursor (EC 1.-.-.-). [Soybean] {Glycine max}, partial
(34%)
Length = 340
Score = 157 bits (398), Expect = 5e-39
Identities = 79/112 (70%), Positives = 89/112 (78%)
Frame = -1
Query: 222 LVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIA 281
L+ T QGV F+AFFV LL PK AHR VG+ EEEA SYT++L IESG +ENV APAIA
Sbjct: 337 LIFTAQGVVFDAFFVWDLLWPKSAHRFVGHPEEEAGISYTQHLNAIESGKVENVRAPAIA 158
Query: 282 IDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGY 333
ID WRLPKDATLK+ +TVI EAHHRDVNHFA DIH GKEL+EAPAP+GY
Sbjct: 157 IDGWRLPKDATLKEEVTVICGGEAHHRDVNHFACDIHHQGKELKEAPAPIGY 2
>TC218177 similar to PIR|T52422|T52422 alternative oxidase-related protein
IMMUTANS [validated] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (70%)
Length = 1084
Score = 76.3 bits (186), Expect = 2e-14
Identities = 49/180 (27%), Positives = 86/180 (47%), Gaps = 20/180 (11%)
Frame = +3
Query: 148 DVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLM 207
D + R+ R +LET+A VP +LH+ + + ++K E+ NE HL+
Sbjct: 411 DTLYHDRHYARFFVLETIARVPYFAFMSVLHMYESFGWWRRADYLKVHFAESWNEMHHLL 590
Query: 208 TMVEL-VQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKD 266
M EL W++RFL + ++ ++Y +SP++A+ +E A +Y +++ +
Sbjct: 591 IMEELGGNAWWFDRFLAQHIAIFYYIMTVLMYAVSPRMAYHFSECVESHAFETYDKFI-E 767
Query: 267 IESGAIENVPAPAIAIDYW-------------------RLPKDATLKDVITVIRADEAHH 307
++ ++ +PAP IA++Y+ R PK L DV IR DEA H
Sbjct: 768 VQGDELKKMPAPEIAVNYYTGDDLYLFDEFQTSRVPNSRRPKIENLYDVFVNIRDDEAEH 947
>TC234838 UP|AOX2_SOYBN (Q41266) Alternative oxidase 2, mitochondrial
precursor , partial (6%)
Length = 725
Score = 44.7 bits (104), Expect = 6e-05
Identities = 18/22 (81%), Positives = 19/22 (85%)
Frame = +3
Query: 313 FASDIHFHGKELREAPAPLGYH 334
F DIHF GKELREAPAP+GYH
Sbjct: 519 FMQDIHFQGKELREAPAPIGYH 584
>TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|00650
CRAL/TRIO phosphatidyl-inositol-transfer protein domain.
ESTs gb|T76582, gb|N06574 and gb|Z25700 come from this
gene. {Arabidopsis thaliana;} , partial (48%)
Length = 1214
Score = 31.6 bits (70), Expect = 0.53
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 4/53 (7%)
Frame = +2
Query: 54 QAAPEEEKK----EEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTE 102
+A PE E+K EEK E+E R E ++ND S+ WG++ +G E
Sbjct: 371 EALPENEEKKNEGEEKEEEEEKRVEVEENDVSI-----WGVTLLPSKGAEGVE 514
>BF070323
Length = 405
Score = 31.6 bits (70), Expect = 0.53
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +3
Query: 305 AHHRDVNHFASDIHFHGKELREAPAPLGY 333
AHH DV H A +IH+ ++ +A P+GY
Sbjct: 3 AHHLDVYHCAYNIHYLWQKYPQAVVPIGY 89
>TC234610 UP|Q9M6B9 (Q9M6B9) Isoflavone synthase 1 (Fragment), partial (19%)
Length = 523
Score = 30.0 bits (66), Expect = 1.5
Identities = 19/56 (33%), Positives = 31/56 (54%)
Frame = +3
Query: 186 QQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLS 241
+ SG ++ LLE AE+E M + E ++ FL+L+ FF++ +VL L S
Sbjct: 171 EASGVFLDTLLEFAEDETMEIKITKEQIKGLVVVSFLLLSYFITFFHSSYVLALNS 338
>TC229563
Length = 669
Score = 29.6 bits (65), Expect = 2.0
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 6/47 (12%)
Frame = +1
Query: 57 PEEEKKEEKAEK------ESLRTEAKKNDGSVVVSSYWGISRPKITR 97
PEE KKEE E E + EAKK + +VVV Y I + R
Sbjct: 163 PEEAKKEEGGENKEEKGGEESKEEAKKEENNVVVVPYNNIDDEGLKR 303
>BE803763 weakly similar to SP|Q41266|AOX2 Alternative oxidase 2
mitochondrial precursor (EC 1.-.-.-). [Soybean] {Glycine
max}, partial (32%)
Length = 325
Score = 29.3 bits (64), Expect = 2.6
Identities = 33/118 (27%), Positives = 52/118 (43%), Gaps = 9/118 (7%)
Frame = +2
Query: 2 KHLALSYALRRAL----NCNRHGLTAVRQLPATEVRRFLVSGE--NGVFSCWNRMMSSQA 55
K AL+ +RR+L N N + L + +P + L+ N F N + S A
Sbjct: 2 KLTALNSTVRRSLLNGLNQNCYHLCSSALMPYSAA*TMLLCSRCTN*WFFYCNMTIVSPA 181
Query: 56 A---PEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMP 110
P +K EE A +++ S+Y+ IS+P + +D TE P N F+P
Sbjct: 182 KAKLPYNDKDEENANAYK----------TIMKSTYYRISKPNVFIDD*TESP*NYFIP 325
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 28.9 bits (63), Expect = 3.4
Identities = 15/39 (38%), Positives = 19/39 (48%)
Frame = -1
Query: 217 WYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEE 255
WY R L G FFN F +Y +P H YL+E+
Sbjct: 410 WYCRILT*RSDGFFFNGFKCVY-YNPGTTHHGFRYLKED 297
>TC210535 similar to GB|AAR28019.1|39545912|AY463617 TAF14b {Arabidopsis
thaliana;} , partial (49%)
Length = 995
Score = 28.9 bits (63), Expect = 3.4
Identities = 16/75 (21%), Positives = 33/75 (43%), Gaps = 4/75 (5%)
Frame = -1
Query: 47 WNRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSS----YWGISRPKITREDGTE 102
W + S P++ KE + ++ + ++ +D + + +S K ++ +
Sbjct: 656 WYHLSSFYLHPQQVWKEAR*VQQVILASSQLDDFAHELKKPQKDQGKLSHHKTPQQQASL 477
Query: 103 WPWNCFMPWETYRPD 117
W WNCF P +T D
Sbjct: 476 WTWNCFHPLDTTSSD 432
>TC220052 similar to GB|AAM51598.1|21436041|AY116964 AT3g47850/T23J7_180
{Arabidopsis thaliana;} , partial (26%)
Length = 855
Score = 28.5 bits (62), Expect = 4.4
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +2
Query: 55 AAPEEEKKEEKAEKESLRTEAKK 77
AAPEEE+ E AE E L E KK
Sbjct: 539 AAPEEEQTEPVAENEDLLNEKKK 607
>TC210742 similar to GB|AAM78055.1|21928053|AY125545 At4g29227 {Arabidopsis
thaliana;} , partial (16%)
Length = 888
Score = 28.5 bits (62), Expect = 4.4
Identities = 14/45 (31%), Positives = 20/45 (44%)
Frame = +2
Query: 88 WGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFL 132
WGI + R G W W C W + +L +T+ +V FL
Sbjct: 236 WGI*KDPTIRAYGYNWCWGCIC-WSCFICNLCQHITRENVAIGFL 367
>TC224556 weakly similar to UP|Q9LWR5 (Q9LWR5) ESTs AU069293(C53946), partial
(52%)
Length = 730
Score = 27.7 bits (60), Expect = 7.6
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +2
Query: 52 SSQAAPEEEKKEEKAEKES 70
++ AAP EKKEEK E+ES
Sbjct: 368 AADAAPAAEKKEEKVEEES 424
>TC229463 weakly similar to UP|O81329 (O81329) F3D13.5 protein, partial (66%)
Length = 1212
Score = 27.7 bits (60), Expect = 7.6
Identities = 14/44 (31%), Positives = 20/44 (44%)
Frame = +1
Query: 126 HVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVP 169
H + D YRT+KLL +P V F C +M + +P
Sbjct: 652 HPAMSIYDYHFYRTMKLLPLPCFVLFLSSIQCNVIMCNLLLLLP 783
>TC215669 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, partial (54%)
Length = 1305
Score = 27.7 bits (60), Expect = 7.6
Identities = 12/36 (33%), Positives = 21/36 (58%)
Frame = +2
Query: 50 MMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVS 85
++S P+EEKK ++ EK E KK + +V++
Sbjct: 500 LLSPIPKPQEEKKVQEEEKPKPTPEEKKEEAQIVMT 607
>TC224580 similar to UP|Q6Y1D6 (Q6Y1D6) 60S acidic ribosomal protein P3
(Fragment), partial (92%)
Length = 642
Score = 27.7 bits (60), Expect = 7.6
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +2
Query: 52 SSQAAPEEEKKEEKAEKES 70
++ AAP EKKEEK E+ES
Sbjct: 365 AADAAPAAEKKEEKVEEES 421
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,348,861
Number of Sequences: 63676
Number of extensions: 200515
Number of successful extensions: 1289
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1279
length of query: 334
length of database: 12,639,632
effective HSP length: 98
effective length of query: 236
effective length of database: 6,399,384
effective search space: 1510254624
effective search space used: 1510254624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0182.2


