

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.18
(485 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
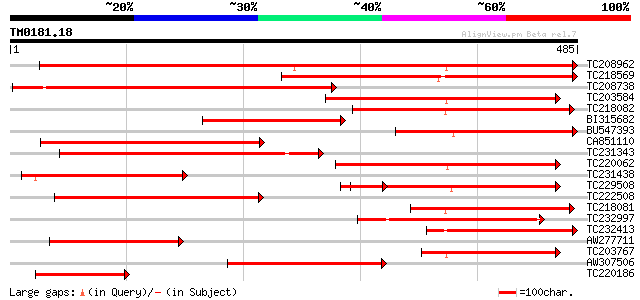
Score E
Sequences producing significant alignments: (bits) Value
TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete 692 0.0
TC218569 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3,... 375 e-104
TC208738 similar to UP|Q7Y076 (Q7Y076) Amino acid permease 6, pa... 340 6e-94
TC203584 weakly similar to UP|Q93X15 (Q93X15) Amino acid permeas... 261 4e-70
TC218082 similar to UP|P92934 (P92934) Amino acid permease 6, pa... 254 5e-68
BI315682 similar to GP|6539602|gb| amino acid transporter b {Vic... 224 5e-59
BU547393 weakly similar to GP|4164408|emb amino acid carrier {Ri... 219 3e-57
CA851110 similar to GP|6539604|gb| amino acid transporter c {Vic... 217 8e-57
TC231343 similar to UP|Q9FF99 (Q9FF99) Amino acid transporter, p... 215 4e-56
TC220062 weakly similar to UP|Q93X13 (Q93X13) Amino acid permeas... 207 8e-54
TC231438 similar to UP|Q93X15 (Q93X15) Amino acid permease AAP1,... 193 2e-49
TC229508 similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15, parti... 183 1e-46
TC222508 weakly similar to UP|Q8GZV4 (Q8GZV4) Amino acid transpo... 175 5e-44
TC218081 weakly similar to UP|P92934 (P92934) Amino acid permeas... 172 4e-43
TC232997 weakly similar to UP|Q9ARG2 (Q9ARG2) Amino acid transpo... 167 1e-41
TC232413 weakly similar to UP|Q93X15 (Q93X15) Amino acid permeas... 159 3e-39
AW277711 147 8e-36
TC203767 weakly similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15... 129 2e-30
AW307506 weakly similar to PIR|T09840|T09 amino acid transport p... 128 7e-30
TC220186 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3,... 127 1e-29
>TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete
Length = 1789
Score = 692 bits (1786), Expect = 0.0
Identities = 323/468 (69%), Positives = 396/468 (84%), Gaps = 8/468 (1%)
Frame = +1
Query: 26 QQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSL 85
Q SKCFDDDGRLKRTGT W A+AHIITAVIGSGVLSLAWA+AQLGWVAGP VM LF++
Sbjct: 136 QSNYSKCFDDDGRLKRTGTFWMATAHIITAVIGSGVLSLAWAVAQLGWVAGPIVMFLFAV 315
Query: 86 VTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTI 145
V YTS LL CYR GD V+G RNYTYM+ V+S +GG +VKLCG+ QY+NLFGVAIGYTI
Sbjct: 316 VNLYTSNLLTQCYRTGDSVSGHRNYTYMEAVNSILGGKKVKLCGLTQYINLFGVAIGYTI 495
Query: 146 ASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAV 205
A+S+SM+AI+RSNC+H S GKDPCHM+ N YMI+FG+ E++ SQIPDFDQ+WWLSI+AA+
Sbjct: 496 AASVSMMAIKRSNCYHSSHGKDPCHMSSNGYMITFGIAEVIFSQIPDFDQVWWLSIVAAI 675
Query: 206 MSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQ------TQKVWRTLQALGDIAFA 259
MSFTYS++GL LG KV EN + GSL GI++GTVTQ TQK+WR+LQALG +AFA
Sbjct: 676 MSFTYSSVGLSLGVAKVAENKSFKGSLMGISIGTVTQAGTVTSTQKIWRSLQALGAMAFA 855
Query: 260 YSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLT 319
YS+S+ILIEIQDT+KSPP+E KTM+KA+ +S+ VTT+FY+LCGC GYAAFGD++PGNLLT
Sbjct: 856 YSFSIILIEIQDTIKSPPAEHKTMRKATTLSIAVTTVFYLLCGCMGYAAFGDNAPGNLLT 1035
Query: 320 GFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTK--TVQIP 377
GFGFYNP+WL+DIAN AIVIHLVGAYQV+SQPLFAFVE ++ K+P S+FVT + IP
Sbjct: 1036GFGFYNPYWLLDIANLAIVIHLVGAYQVFSQPLFAFVEKWSVRKWPKSNFVTAEYDIPIP 1215
Query: 378 LFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQK 437
F Y++N FRLVWRTIFV++TTLI+ML+PFFND+VG+LGA GFWPLTVYFP++MYI QK
Sbjct: 1216CFGVYQLNFFRLVWRTIFVLLTTLIAMLMPFFNDVVGILGAFGFWPLTVYFPIDMYISQK 1395
Query: 438 RIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKAIY 485
+I +W+ +WI QL S +CLI+++ AA GS+AG+ LDL+ YKPFK Y
Sbjct: 1396KIGRWTSRWIGLQLLSASCLIISLLAAVGSMAGVVLDLKTYKPFKTSY 1539
>TC218569 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3, partial
(51%)
Length = 1108
Score = 375 bits (963), Expect = e-104
Identities = 178/255 (69%), Positives = 214/255 (83%), Gaps = 2/255 (0%)
Frame = +3
Query: 233 TGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVV 292
+GI++GTVT+ QKVW QALG+IAFAYSYS +L+EIQDT+KSPPSE KTMKKA+ +S+
Sbjct: 6 SGISIGTVTEAQKVWGVFQALGNIAFAYSYSFVLLEIQDTIKSPPSEVKTMKKAAKLSIA 185
Query: 293 VTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPL 352
VTT FYMLCGC GYAAFGDS+PGNLL GFGF+ +WLIDIANAAIVIHLVGAYQVY+QPL
Sbjct: 186 VTTTFYMLCGCVGYAAFGDSAPGNLLAGFGFHKLYWLIDIANAAIVIHLVGAYQVYAQPL 365
Query: 353 FAFVENYAAEKFP--DSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFN 410
FAFVE A+++P D +F + IP +Y N+F LVWRT+FVIITT+ISMLLPFFN
Sbjct: 366 FAFVEKETAKRWPKIDKEF---QISIPGLQSYNQNIFSLVWRTVFVIITTVISMLLPFFN 536
Query: 411 DIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAG 470
DI+G++GALGFWPLTVYFPVEMYI+QKRIPKWS++WI +L S+ CLIVT+AA GS+ G
Sbjct: 537 DILGVIGALGFWPLTVYFPVEMYILQKRIPKWSMRWISLELLSVVCLIVTIAAGLGSMVG 716
Query: 471 IFLDLQVYKPFKAIY 485
+ LDLQ YKPF + Y
Sbjct: 717 VLLDLQKYKPFSSDY 761
>TC208738 similar to UP|Q7Y076 (Q7Y076) Amino acid permease 6, partial (52%)
Length = 966
Score = 340 bits (873), Expect = 6e-94
Identities = 172/279 (61%), Positives = 214/279 (76%), Gaps = 2/279 (0%)
Frame = +3
Query: 3 MNEKNGSKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVL 62
++ +N N+ Q + ++ + GG K FDDDGR++RTGT TASAHIITAVIGSGVL
Sbjct: 132 LSNRN*EMNSDQFQKNSMFVETPEDGG-KNFDDDGRVRRTGTWITASAHIITAVIGSGVL 308
Query: 63 SLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGG 122
SLAWAIAQ+GWVAGPAV+ FS +TY+TS LL CYR+ DPV+GKRNYTY DVV S +GG
Sbjct: 309 SLAWAIAQMGWVAGPAVLFAFSFITYFTSTLLADCYRSPDPVHGKRNYTYSDVVRSVLGG 488
Query: 123 IQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGL 182
+ +LCG+ QY+NL GV IGYTI +SISM+A++RSNCFHK D C+ + N +MI F
Sbjct: 489 RKFQLCGLAQYINLVGVTIGYTITASISMVAVKRSNCFHKHGHHDKCYTSNNPFMILFAC 668
Query: 183 VEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENG-AVGGSLTGITVGT-V 240
++IVLSQIP+F +LWWLSI+AAVMSF YS+IGLGL KV G V +LTG+ VG V
Sbjct: 669 IQIVLSQIPNFHKLWWLSIVAAVMSFAYSSIGLGLSVAKVAGGGEPVRTTLTGVQVGVDV 848
Query: 241 TQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSE 279
T ++KVWRT QA+GDIAFAY+YS +LIEIQDT+KS P E
Sbjct: 849 TGSEKVWRTFQAIGDIAFAYAYSNVLIEIQDTLKSSPPE 965
>TC203584 weakly similar to UP|Q93X15 (Q93X15) Amino acid permease AAP1,
partial (34%)
Length = 953
Score = 261 bits (668), Expect = 4e-70
Identities = 113/203 (55%), Positives = 157/203 (76%), Gaps = 2/203 (0%)
Frame = +2
Query: 271 DTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLI 330
DT++SPP E++TMKKAS +++ +TT FY+ CGCFGYAAFG+ +PGNLLTGFGF+ PFWLI
Sbjct: 2 DTLESPPPENQTMKKASMVAIFITTFFYLCCGCFGYAAFGNDTPGNLLTGFGFFEPFWLI 181
Query: 331 DIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTK--TVQIPLFNAYKINLFR 388
D+ANA I++HLVG YQ+YSQP+++ V+ +A+ KFP+S FV V++PL +++NLFR
Sbjct: 182 DLANACIILHLVGGYQIYSQPIYSTVDRWASRKFPNSGFVNNFYRVKLPLLPGFQLNLFR 361
Query: 389 LVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWIC 448
+RT +VI T +++ P+FN I+G+LGA+ FWPL +YFPVEMY +Q++I WS KWI
Sbjct: 362 FCFRTTYVISTIGLAIFFPYFNQILGVLGAINFWPLAIYFPVEMYFVQQKIAAWSSKWIV 541
Query: 449 FQLFSLACLIVTVAAAAGSVAGI 471
+ FS AC +VTV GS+ GI
Sbjct: 542 LRTFSFACFLVTVMGLVGSLEGI 610
>TC218082 similar to UP|P92934 (P92934) Amino acid permease 6, partial (40%)
Length = 638
Score = 254 bits (650), Expect = 5e-68
Identities = 113/192 (58%), Positives = 147/192 (75%), Gaps = 2/192 (1%)
Frame = +1
Query: 294 TTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLF 353
TTLFY+LCGC GYAAFG+ +PGN LTGFGFY PFWLID AN I +HLVGAYQV+ QP+F
Sbjct: 4 TTLFYVLCGCLGYAAFGNDAPGNFLTGFGFYEPFWLIDFANICIAVHLVGAYQVFCQPIF 183
Query: 354 AFVENYAAEKFPDSDFVT--KTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFND 411
FVEN+ E++P+S FV + PL + +N FR+VWRT +VIIT LI+M+ PFFND
Sbjct: 184 GFVENWGKERWPNSQFVNGEHALNFPLCGTFPVNFFRVVWRTTYVIITALIAMMFPFFND 363
Query: 412 IVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGI 471
+GL+G+L FWPLTVYFP+EMYI Q ++ ++S W ++ S ACLIV++ +AAGS+ G+
Sbjct: 364 FLGLIGSLSFWPLTVYFPIEMYIKQSKMQRFSFTWTWLKILSWACLIVSIISAAGSIQGL 543
Query: 472 FLDLQVYKPFKA 483
DL+ Y+PFKA
Sbjct: 544 AQDLKKYQPFKA 579
>BI315682 similar to GP|6539602|gb| amino acid transporter b {Vicia faba},
partial (46%)
Length = 367
Score = 224 bits (572), Expect = 5e-59
Identities = 108/122 (88%), Positives = 116/122 (94%)
Frame = +1
Query: 166 KDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIEN 225
KDPCH+N N+YMISFG+VEI+ SQIP FDQLWWLSI+AAVMSFTYSTIGLGLG GKVIEN
Sbjct: 1 KDPCHINSNMYMISFGIVEILFSQIPGFDQLWWLSIVAAVMSFTYSTIGLGLGIGKVIEN 180
Query: 226 GAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKK 285
G VGGSLTGIT+GTVTQT KVWRT+QALGDIAFAYSYS+ILIEIQDTVKSPPSESKTMKK
Sbjct: 181 GGVGGSLTGITIGTVTQTDKVWRTMQALGDIAFAYSYSLILIEIQDTVKSPPSESKTMKK 360
Query: 286 AS 287
AS
Sbjct: 361 AS 366
>BU547393 weakly similar to GP|4164408|emb amino acid carrier {Ricinus
communis}, partial (33%)
Length = 678
Score = 219 bits (557), Expect = 3e-57
Identities = 102/157 (64%), Positives = 128/157 (80%), Gaps = 2/157 (1%)
Frame = -3
Query: 331 DIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPL--FNAYKINLFR 388
DIANA IVIHLVGAYQVY QP FVE+ AAE+F +SDF+++ ++P+ K+NLFR
Sbjct: 676 DIANAGIVIHLVGAYQVYCQPXXXFVESNAAERFXNSDFMSREFEVPIPGCKXXKLNLFR 497
Query: 389 LVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWIC 448
LVWRT+FVI++T+I+MLLPFFNDIVGL+GA+GFWPLTVY PVEMYI Q +IPKW IKWI
Sbjct: 496 LVWRTLFVILSTVIAMLLPFFNDIVGLIGAIGFWPLTVYLPVEMYITQTKIPKWGIKWIG 317
Query: 449 FQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKAIY 485
Q+ S+AC ++T+ AAAGS+AG+ DL+VYKPF Y
Sbjct: 316 LQMLSVACFVITILAAAGSIAGVIDDLKVYKPFVTSY 206
>CA851110 similar to GP|6539604|gb| amino acid transporter c {Vicia faba},
partial (65%)
Length = 698
Score = 217 bits (553), Expect = 8e-57
Identities = 103/192 (53%), Positives = 137/192 (70%)
Frame = +1
Query: 27 QGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLV 86
+ G +DDDG KRTG + +A AHIITAVIGSGVLSLAW+ +QLGW+ GP ++ ++V
Sbjct: 118 RSGIGAYDDDGHAKRTGNLQSAVAHIITAVIGSGVLSLAWSTSQLGWIGGPFSLLCCAIV 297
Query: 87 TYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIA 146
TY +S LL CYR DPV GKRNY+YMD V +G + + G +Q+L L+G +I Y +
Sbjct: 298 TYISSFLLSDCYRTPDPVTGKRNYSYMDAVRVYLGYKRTCVAGFLQFLTLYGTSIAYVLT 477
Query: 147 SSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVM 206
++ S+ AI RSNC+HK + PC GN+YM FGLV+IV+S IPD + W+S++AA+M
Sbjct: 478 TATSLSAILRSNCYHKKGHEAPCKYGGNLYMALFGLVQIVMSFIPDLHNMAWVSVVAALM 657
Query: 207 SFTYSTIGLGLG 218
SFTYS IGLGLG
Sbjct: 658 SFTYSFIGLGLG 693
>TC231343 similar to UP|Q9FF99 (Q9FF99) Amino acid transporter, partial (27%)
Length = 700
Score = 215 bits (547), Expect = 4e-56
Identities = 108/227 (47%), Positives = 154/227 (67%), Gaps = 1/227 (0%)
Frame = +3
Query: 43 GTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGD 102
GTVWTA AHI+T VIGSGVLSL W+ AQLGW+AGP ++L + T ++S LLC YR+
Sbjct: 24 GTVWTAVAHIVTGVIGSGVLSLPWSTAQLGWLAGPFSILLIASTTLFSSFLLCNTYRHPH 203
Query: 103 PVNG-KRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFH 161
P G R+ +Y+DVVH ++G +L G++ ++L+G AI + I ++IS+ I+ S C+H
Sbjct: 204 PEYGPNRSASYLDVVHLHLGISNGRLSGLLVSISLYGFAIAFVITTAISLRTIQNSFCYH 383
Query: 162 KSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGK 221
+ C YM+ FG ++IVLSQIP+F + WLS++AA+MSFTYS IG+GL +
Sbjct: 384 NKGPEAACESVDAYYMLLFGAIQIVLSQIPNFHNIKWLSVVAAIMSFTYSFIGMGLSIAQ 563
Query: 222 VIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIE 268
+IE G GS+ GI+ T +K+W QALGDI+F+Y +S IL+E
Sbjct: 564 IIEKGHAEGSIGGIS--TSNGAEKLWLVSQALGDISFSYPFSTILME 698
>TC220062 weakly similar to UP|Q93X13 (Q93X13) Amino acid permease AAP4,
partial (29%)
Length = 818
Score = 207 bits (527), Expect = 8e-54
Identities = 98/195 (50%), Positives = 136/195 (69%), Gaps = 2/195 (1%)
Frame = +2
Query: 279 ESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIV 338
E++TMKKAS I+V VTT Y CG YAAFGD++PGNLLTGF +WL++ ANA IV
Sbjct: 5 ENQTMKKASVIAVSVTTFLYXSCGGXXYAAFGDNTPGNLLTGFVSSKSYWLVNFANACIV 184
Query: 339 IHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKT--VQIPLFNAYKINLFRLVWRTIFV 396
+HLVG+YQVYSQPLF VEN+ +FPDS+FV T +++PL A+++N L +RT +V
Sbjct: 185 VHLVGSYQVYSQPLFGTVENWFRFRFPDSEFVNHTYILKLPLLPAFELNFLSLSFRTAYV 364
Query: 397 IITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLAC 456
TT+I+M+ P+FN I+G+LG++ FWPLT+YFPVE+Y+ Q W+ KW+ + FS
Sbjct: 365 ASTTVIAMIFPYFNQILGVLGSIIFWPLTIYFPVEIYLSQSSTVSWTTKWVLLRTFSFFG 544
Query: 457 LIVTVAAAAGSVAGI 471
+ + G + GI
Sbjct: 545 FLFGLFTLIGCIKGI 589
>TC231438 similar to UP|Q93X15 (Q93X15) Amino acid permease AAP1, partial
(26%)
Length = 659
Score = 193 bits (490), Expect = 2e-49
Identities = 95/146 (65%), Positives = 113/146 (77%), Gaps = 4/146 (2%)
Frame = +1
Query: 11 NNHHHQAFDV---SLD-MQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAW 66
N HHQ F + S+D + Q SK +DDDG +KRTGTVWT S+HIITAV+GSGVLSLAW
Sbjct: 130 NLSHHQDFGMEPYSIDGVSSQTNSKFYDDDGHVKRTGTVWTTSSHIITAVVGSGVLSLAW 309
Query: 67 AIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVK 126
A+AQ+GWVAGP VMI FS+VT YT+ LL CYR GDPV GKRNYT+MD V S +GG
Sbjct: 310 AMAQMGWVAGPVVMIFFSVVTLYTTSLLADCYRCGDPVTGKRNYTFMDAVQSILGGYYDT 489
Query: 127 LCGIVQYLNLFGVAIGYTIASSISMI 152
CG+VQY NL+G A+GYTIA+SISM+
Sbjct: 490 FCGVVQYSNLYGTAVGYTIAASISMM 567
>TC229508 similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15, partial (39%)
Length = 865
Score = 183 bits (465), Expect = 1e-46
Identities = 82/182 (45%), Positives = 124/182 (68%), Gaps = 2/182 (1%)
Frame = +1
Query: 292 VVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQP 351
++ L +ML C F S+ L FGFY P+WLID ANA IV+HLVG YQ+YSQP
Sbjct: 37 ILLPLLWMLWIC----GFWKSNTRKPLDRFGFYEPYWLIDFANACIVLHLVGGYQIYSQP 204
Query: 352 LFAFVENYAAEKFPDSDFVTKTVQI--PLFNAYKINLFRLVWRTIFVIITTLISMLLPFF 409
++ V+ + ++++P+S FV Q+ P A+++N+FR+ +RT +V+ TT +++L P+F
Sbjct: 205 IYGAVDRWCSKRYPNSGFVNNFYQLKLPRLPAFQLNMFRICFRTAYVVSTTGLAILFPYF 384
Query: 410 NDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVA 469
N ++G+LGALGFWPL +YFPVEMY +Q+++ WS KWI + FS C +V++ GS+
Sbjct: 385 NQVIGVLGALGFWPLAIYFPVEMYFVQRKVEAWSRKWIVLRTFSFICFLVSLLGLIGSLE 564
Query: 470 GI 471
GI
Sbjct: 565 GI 570
Score = 67.0 bits (162), Expect = 2e-11
Identities = 27/40 (67%), Positives = 34/40 (84%)
Frame = +2
Query: 284 KKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGF 323
KKAS I++++TT FY+ CGCFGYAAFG+ +PGNLLTG F
Sbjct: 2 KKASMIAILITTFFYLCCGCFGYAAFGNQTPGNLLTGLDF 121
>TC222508 weakly similar to UP|Q8GZV4 (Q8GZV4) Amino acid transporter,
partial (22%)
Length = 700
Score = 175 bits (443), Expect = 5e-44
Identities = 84/180 (46%), Positives = 121/180 (66%), Gaps = 1/180 (0%)
Frame = +1
Query: 39 LKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACY 98
++RTGTVWTA AHI+T VIGSGVLSLAW+IAQLGW+ GP ++ F+ +T +S LL Y
Sbjct: 154 VERTGTVWTAVAHIVTGVIGSGVLSLAWSIAQLGWIGGPLTIVFFAAITLLSSFLLSNTY 333
Query: 99 RNGDPVNGK-RNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERS 157
R+ DP G R+ +Y+D V+ + G C + ++L+G I Y I ++ISM AI++S
Sbjct: 334 RSPDPELGPHRSSSYLDAVNLHKGEGNSHFCAVFVNVSLYGFGIAYVITAAISMRAIQKS 513
Query: 158 NCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGL 217
NC + + C +M+ FG ++++LSQIP+F + WLSILA +MSF Y+ IG+GL
Sbjct: 514 NCSQDNGNEXTCGFGDGYFMLIFGAMQVLLSQIPNFHNIQWLSILAXIMSFAYAFIGMGL 693
>TC218081 weakly similar to UP|P92934 (P92934) Amino acid permease 6, partial
(28%)
Length = 636
Score = 172 bits (435), Expect = 4e-43
Identities = 77/142 (54%), Positives = 107/142 (75%), Gaps = 2/142 (1%)
Frame = +3
Query: 344 AYQVYSQPLFAFVENYAAEKFPDSDFVT--KTVQIPLFNAYKINLFRLVWRTIFVIITTL 401
AYQV+ QP+F FVEN+ E++P+S FV ++ PLF + +N FR+VWRT +VIIT L
Sbjct: 9 AYQVFCQPIFGFVENWGKERWPNSHFVNGEHALKFPLFGTFPVNFFRVVWRTTYVIITAL 188
Query: 402 ISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTV 461
I+M+ PFFND +GL+G+L FWPLTVYFP+EMYI Q ++ K+S W ++ S ACLIV++
Sbjct: 189 IAMMFPFFNDFLGLIGSLSFWPLTVYFPIEMYIKQSKMQKFSFTWTWLKILSWACLIVSI 368
Query: 462 AAAAGSVAGIFLDLQVYKPFKA 483
+AAGS+ G+ DL+ Y+PFKA
Sbjct: 369 ISAAGSIQGLAQDLKKYQPFKA 434
>TC232997 weakly similar to UP|Q9ARG2 (Q9ARG2) Amino acid transporter,
partial (18%)
Length = 487
Score = 167 bits (422), Expect = 1e-41
Identities = 81/161 (50%), Positives = 113/161 (69%), Gaps = 1/161 (0%)
Frame = +3
Query: 298 YMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVE 357
++LC GYAAFGD++PGN+LTGF PFWL+ + N IVIH++GAYQV QP F VE
Sbjct: 6 FLLCSGLGYAAFGDNTPGNILTGFT--EPFWLVALGNGFIVIHMIGAYQVMGQPFFRIVE 179
Query: 358 NYAAEKFPDSDFVTKTVQIPLFNAY-KINLFRLVWRTIFVIITTLISMLLPFFNDIVGLL 416
A +P+SDF+ K + + NLFRLVWRTIFVI+ T+++M++PFF++++ LL
Sbjct: 180 IGANIAWPNSDFINKEYPFIVGGLMVRFNLFRLVWRTIFVILATILAMVMPFFSEVLSLL 359
Query: 417 GALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACL 457
GA+GF PL V+ P++M+I QK I K S++W Q L+CL
Sbjct: 360 GAIGFGPLVVFIPIQMHIAQKSIRKLSLRWCGLQF--LSCL 476
>TC232413 weakly similar to UP|Q93X15 (Q93X15) Amino acid permease AAP1,
partial (27%)
Length = 802
Score = 159 bits (402), Expect = 3e-39
Identities = 75/129 (58%), Positives = 100/129 (77%)
Frame = +3
Query: 357 ENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLL 416
E +A++++P+ + K + IP F+ Y ++ FRLVWRT+FVIITT ++ML+PFFND++GLL
Sbjct: 3 EKWASKRWPEVETEYK-IPIPGFSPYNLSPFRLVWRTVFVIITTFVAMLIPFFNDVLGLL 179
Query: 417 GALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQ 476
GALGFWPL+V+ PV+M I QKR P+WS +WI Q+ S+ C IV+VAAA GSVA I LDLQ
Sbjct: 180 GALGFWPLSVFLPVQMSIKQKRTPRWSGRWIGMQILSVVCFIVSVAAAVGSVASIVLDLQ 359
Query: 477 VYKPFKAIY 485
YKPF Y
Sbjct: 360 KYKPFHVDY 386
>AW277711
Length = 346
Score = 147 bits (372), Expect = 8e-36
Identities = 69/114 (60%), Positives = 84/114 (73%)
Frame = +3
Query: 35 DDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILL 94
DDGR++RTG VWTAS HIIT V+G+GVLSLAW +AQLGW+AG A +I FS V+ +T L+
Sbjct: 3 DDGRIRRTGNVWTASIHIITVVVGAGVLSLAWVMAQLGWLAGIASIITFSAVSIFTYNLV 182
Query: 95 CACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASS 148
CYR DPV GKRNYTYM V + +GG CG+VQY L G+ +GYTI SS
Sbjct: 183 ADCYRYPDPVTGKRNYTYMQAVKAYLGGTMHVFCGLVQYTKLAGITVGYTITSS 344
>TC203767 weakly similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15, partial
(30%)
Length = 635
Score = 129 bits (325), Expect = 2e-30
Identities = 58/121 (47%), Positives = 84/121 (68%), Gaps = 2/121 (1%)
Frame = +3
Query: 353 FAFVENYAAEKFPDSDFVTK--TVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFN 410
++ V+ +A+ KFP+S FV V++PL +++NLFR +RT +VI TT +++ P+FN
Sbjct: 3 YSTVDRWASRKFPNSGFVNNFYKVKLPLLPGFQLNLFRFCFRTTYVISTTGLAIFFPYFN 182
Query: 411 DIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAG 470
I+G+LGA+ FWPL +YFPVEMY +Q +I WS KWI + FS AC +VTV GS+ G
Sbjct: 183 PILGVLGAINFWPLAIYFPVEMYFVQNKIAAWSSKWIVLRTFSFACFLVTVMGLVGSLEG 362
Query: 471 I 471
I
Sbjct: 363 I 365
>AW307506 weakly similar to PIR|T09840|T09 amino acid transport protein AAP2
- castor bean (fragment), partial (40%)
Length = 413
Score = 128 bits (321), Expect = 7e-30
Identities = 64/137 (46%), Positives = 89/137 (64%), Gaps = 1/137 (0%)
Frame = +3
Query: 187 LSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGT-VTQTQK 245
LSQIP+F +L LS +AA+ SF Y+ IG GL V+ + G VG +++ K
Sbjct: 3 LSQIPNFHKLTCLSTVAAITSFCYALIGSGLSLAVVVSGKGETTRVFGNKVGPGLSEADK 182
Query: 246 VWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFG 305
+WR ALG+IA A SY+ ++ +I DT+KS P E K MKKA+ + + T+ ++LCG G
Sbjct: 183 MWRVFSALGNIALACSYATVVYDIMDTLKSYPPECKQMKKANVLGITTMTILFLLCGSLG 362
Query: 306 YAAFGDSSPGNLLTGFG 322
YAAFGD +PGN+LTGFG
Sbjct: 363 YAAFGDDTPGNILTGFG 413
>TC220186 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3, partial
(21%)
Length = 604
Score = 127 bits (318), Expect = 1e-29
Identities = 57/80 (71%), Positives = 66/80 (82%)
Frame = +1
Query: 23 DMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMIL 82
DM + +C+DDDG LKRTGT+WT S+HIITAV+GSGVLSLAWAIAQ+GW+AGPAVMIL
Sbjct: 364 DMPLKSDPECYDDDGHLKRTGTIWTTSSHIITAVVGSGVLSLAWAIAQMGWIAGPAVMIL 543
Query: 83 FSLVTYYTSILLCACYRNGD 102
FS+VT YTS L CY GD
Sbjct: 544 FSIVTLYTSSFLADCYHTGD 603
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,074,734
Number of Sequences: 63676
Number of extensions: 354629
Number of successful extensions: 2715
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 2651
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2684
length of query: 485
length of database: 12,639,632
effective HSP length: 101
effective length of query: 384
effective length of database: 6,208,356
effective search space: 2384008704
effective search space used: 2384008704
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0181.18


