

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.7
(199 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
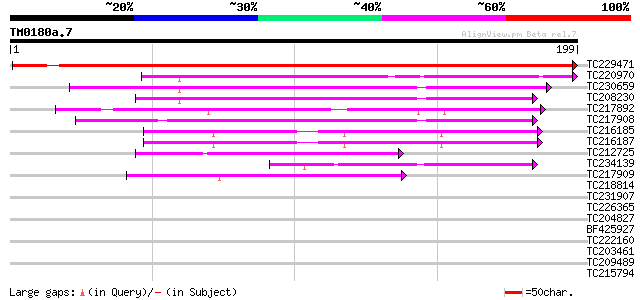
Score E
Sequences producing significant alignments: (bits) Value
TC229471 UP|Q8V7C6 (Q8V7C6) ORF1 (Fragment), partial (19%) 323 2e-89
TC220970 63 1e-10
TC230659 weakly similar to UP|O24088 (O24088) MtN24 protein, par... 58 3e-09
TC208230 similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (9%) 52 2e-07
TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial... 52 2e-07
TC217908 50 9e-07
TC216185 49 2e-06
TC216187 47 8e-06
TC212725 44 4e-05
TC234139 similar to UP|O24088 (O24088) MtN24 protein, partial (22%) 42 2e-04
TC217909 41 4e-04
TC218814 35 0.023
TC231907 32 0.25
TC226365 weakly similar to UP|Q944E4 (Q944E4) Cellulose synthase... 30 0.73
TC204827 UP|Q9SLQ6 (Q9SLQ6) Actin isoform B, complete 29 1.6
BF425927 28 2.8
TC222160 28 2.8
TC203461 28 2.8
TC209489 27 6.2
TC215794 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyru... 27 6.2
>TC229471 UP|Q8V7C6 (Q8V7C6) ORF1 (Fragment), partial (19%)
Length = 905
Score = 323 bits (829), Expect = 2e-89
Identities = 163/198 (82%), Positives = 175/198 (88%)
Frame = +1
Query: 2 MRSPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVF 61
MRSPQPLRNG D N+ S +PRFHST+AEHKLRRFN LILVFRL SFSF LASSVF
Sbjct: 121 MRSPQPLRNGGD----NSCSPSHNPRFHSTVAEHKLRRFNLLILVFRLSSFSFYLASSVF 288
Query: 62 MLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFD 121
MLTNT GS SPHW HYDTFRF LAANAIVAVYS+FEM ASVWEISRG T+FPEVLQ+WFD
Sbjct: 289 MLTNTRGSYSPHWNHYDTFRFFLAANAIVAVYSVFEMGASVWEISRGVTLFPEVLQIWFD 468
Query: 122 FGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSL 181
FGHDQVFAYLLL+ASAAGT+M RTLKE +TC ASNAFCVQ+DIA+ALGYA F+FLGFTSL
Sbjct: 469 FGHDQVFAYLLLSASAAGTSMARTLKEMETCRASNAFCVQTDIAIALGYAAFLFLGFTSL 648
Query: 182 LTGFRVVCFIINGSRFHL 199
LTGFRVV FIINGSRFHL
Sbjct: 649 LTGFRVVSFIINGSRFHL 702
>TC220970
Length = 636
Score = 62.8 bits (151), Expect = 1e-10
Identities = 45/154 (29%), Positives = 73/154 (47%), Gaps = 1/154 (0%)
Frame = +1
Query: 47 FRLISFSFSLAS-SVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
FRL L S SV T G + Y +R+ L+ N I VY+ F+ +++
Sbjct: 100 FRLSEVVLCLISFSVMAADKTRGWSGDSFDRYKEYRYCLSVNVIAFVYAAFQAGDLAYQV 279
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIA 165
G + L+ FDF DQV AYLL+ SAA +A R + + + + F + +
Sbjct: 280 VTGRRIINHHLRYHFDFFMDQVLAYLLI--SAASSAATR-VDDWQSNWGKDEFTEMASAS 450
Query: 166 VALGYAGFVFLGFTSLLTGFRVVCFIINGSRFHL 199
+AL + FV +SL +G+ +C + + F+L
Sbjct: 451 IALAFLAFVAFAISSLFSGYN-LCTLFS*QHFNL 549
>TC230659 weakly similar to UP|O24088 (O24088) MtN24 protein, partial (44%)
Length = 1181
Score = 58.2 bits (139), Expect = 3e-09
Identities = 46/170 (27%), Positives = 76/170 (44%), Gaps = 1/170 (0%)
Frame = +2
Query: 22 RSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLAS-SVFMLTNTHGSDSPHWYHYDTF 80
R P + K+ ++ L+L R+ +F F LAS SV G +Y Y F
Sbjct: 464 RRLRPDMTGLLRSKKIATWSKLLLGLRVTAFVFCLASFSVLATDKKQGWAIDSFYLYKEF 643
Query: 81 RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGT 140
R+ L+ N I V+S ++ + G + L+ +F F DQ+ YLL++AS++
Sbjct: 644 RYSLSVNVIGFVHSALQICDLGRYFATGKHLVEHQLRGYFTFALDQILTYLLMSASSSAA 823
Query: 141 AMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCF 190
+ + + F ++ +VAL FV SL++G VV F
Sbjct: 824 TRA---YDWVSNWGEDKFPYMANASVALSLVAFVAFALASLVSGSIVVRF 964
>TC208230 similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (9%)
Length = 938
Score = 52.0 bits (123), Expect = 2e-07
Identities = 37/142 (26%), Positives = 64/142 (45%), Gaps = 1/142 (0%)
Frame = +2
Query: 45 LVFRLISFSFSLAS-SVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVW 103
L FRL L S SV T G + Y +R+ L+ N I YS +
Sbjct: 242 LGFRLSEVVVCLISFSVMAADKTQGWSGDSFDRYKEYRYCLSVNIIGFAYSALQACDLTC 421
Query: 104 EISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSD 163
+++ G + L+ FDF DQV AYLL+++S++ V ++ + F +
Sbjct: 422 QLATGKRLISHHLRNHFDFFMDQVLAYLLISSSSSAATRV---EDWILNWGKDEFTEMAT 592
Query: 164 IAVALGYAGFVFLGFTSLLTGF 185
++ + + FV +SL++G+
Sbjct: 593 ASIGMAFLAFVAFAISSLISGY 658
>TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial (10%)
Length = 989
Score = 51.6 bits (122), Expect = 2e-07
Identities = 50/185 (27%), Positives = 80/185 (43%), Gaps = 13/185 (7%)
Frame = +1
Query: 17 NNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTHG------SD 70
N N RS+S + H + L F + R+I S AS M+TN
Sbjct: 157 NPNQPRSASQQQHRII----LTIFLMAQDILRIIPTLLSAASIAVMVTNNQTVLIFAIRF 324
Query: 71 SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAY 130
H+Y+ +F+F +AAN +V SL +V + + + + +F F HD V
Sbjct: 325 QAHFYYSPSFKFFVAANGVVVALSLLTLVLNFLMKRQASPRYH-----FFLFLHDIVMMV 489
Query: 131 LLLAASAAGTAM--VRTLKETDT-----CTASNAFCVQSDIAVALGYAGFVFLGFTSLLT 183
LL+A AA TA+ V E C FC + +++ L Y F+ +LL+
Sbjct: 490 LLIAGCAAATAIGYVGQFGEDHVGWQPICDHVRKFCTTNLVSLLLSYFAFISYFGITLLS 669
Query: 184 GFRVV 188
+++V
Sbjct: 670 AYKIV 684
>TC217908
Length = 821
Score = 49.7 bits (117), Expect = 9e-07
Identities = 40/162 (24%), Positives = 73/162 (44%)
Frame = +2
Query: 24 SSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFV 83
++P ST++ L LI VF LI+ + ++ T+ + +R++
Sbjct: 50 NTPPSSSTVSRTVLLLLRVLIFVFLLIAL---ILIAIVKQTDDETGAEIKFNDIYAYRYM 220
Query: 84 LAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMV 143
++ I Y+L +M S++ + G V FDF D++ +YLL++ SAAG +
Sbjct: 221 ISTIIIGFAYNLLQMAFSMFTVVSGNRVLSGDGGYLFDFFGDKIISYLLISGSAAGFGVT 400
Query: 144 RTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
E SN+F +++ + +L GF+ S T F
Sbjct: 401 ---VELGRGVPSNSFIEKANASASLLLIGFLLTAIASTFTSF 517
>TC216185
Length = 1098
Score = 48.9 bits (115), Expect = 2e-06
Identities = 39/150 (26%), Positives = 68/150 (45%), Gaps = 10/150 (6%)
Frame = +1
Query: 48 RLISFSFSLASSVFMLTNTHGSD--SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
RL+ +++ V ML N+ ++ S + FR+++ AN I A YSLF V +
Sbjct: 328 RLVPVGLCVSALVLMLKNSQQNEYGSVDYSDLGAFRYLVHANGICAGYSLFSAVIA---- 495
Query: 106 SRGATVFPEVL-QVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD-------TCTASNA 157
A P + + W F DQV Y++LAA A T ++ + D C +
Sbjct: 496 ---AMPCPSTIPRAWTFFLLDQVLTYIILAAGAVSTEVLYLAENGDAATTWSSACGSFGR 666
Query: 158 FCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
FC + +VA+ + SL++ +++
Sbjct: 667 FCHKVTASVAITFVAVFCYVLLSLVSSYKL 756
>TC216187
Length = 977
Score = 46.6 bits (109), Expect = 8e-06
Identities = 38/150 (25%), Positives = 68/150 (45%), Gaps = 10/150 (6%)
Frame = +1
Query: 48 RLISFSFSLASSVFMLTNTHGSD--SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
RL +++ V ML ++ ++ S + FR+++ AN I A YSLF V +
Sbjct: 229 RLFPVGLCVSALVLMLKSSQQNEYGSVDYSDLGAFRYLVHANGICAGYSLFSAVIA---- 396
Query: 106 SRGATVFPEVL-QVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD-------TCTASNA 157
A P + + W F DQV Y++LAA A T ++ ++ D C +
Sbjct: 397 ---AMPCPSTIPRAWTFFLLDQVLTYIILAAGAVSTEVLYLAEKGDAATTWSSACGSFGR 567
Query: 158 FCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
FC + +VA+ + SL++ +++
Sbjct: 568 FCHKVTASVAITFVAVFCYVLLSLISSYKL 657
>TC212725
Length = 415
Score = 44.3 bits (103), Expect = 4e-05
Identities = 26/94 (27%), Positives = 50/94 (52%)
Frame = +1
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWE 104
L R ++ FSL S M +N HG D + Y+ +R++LA + ++Y+ + + E
Sbjct: 118 LGLRGVALLFSLISFFIMASNKHG-DWREFDKYEEYRYLLAIAILSSLYTGAQAFRLLQE 294
Query: 105 ISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAA 138
+S + + DF DQ+ AYLL++++++
Sbjct: 295 LSTAKQLLQPRMAAMIDFFGDQIIAYLLISSASS 396
>TC234139 similar to UP|O24088 (O24088) MtN24 protein, partial (22%)
Length = 497
Score = 41.6 bits (96), Expect = 2e-04
Identities = 29/95 (30%), Positives = 54/95 (56%), Gaps = 1/95 (1%)
Frame = +2
Query: 92 VYSLFEMVASV-WEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD 150
VYS ++ V + I++ T+ P+ L+ +FDF DQ AYLL++AS++ +V T+
Sbjct: 8 VYSGLQIYDIVKYMITKRHTMDPK-LRGYFDFAMDQALAYLLMSASSSAATIV--CSWTN 178
Query: 151 TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
+ A++ ++ VAL + FV +S+++ F
Sbjct: 179 SVLAADKHVQMANACVALSFVAFVAFASSSVVSAF 283
>TC217909
Length = 445
Score = 40.8 bits (94), Expect = 4e-04
Identities = 25/102 (24%), Positives = 51/102 (49%), Gaps = 4/102 (3%)
Frame = +2
Query: 42 SLILVFRLISFSFSLASSVFMLTNTHGSDSP----HWYHYDTFRFVLAANAIVAVYSLFE 97
+++L+ R+++F F L + + + D + +R++++ I Y+L +
Sbjct: 83 TVLLLLRVLTFVFLLIALILIAIVKQTDDETGVEIKFNDIYAYRYMISTIIIGFAYNLLQ 262
Query: 98 MVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG 139
M S++ + G V FDF D++ +YLL++ SAAG
Sbjct: 263 MALSIFTVVSGNRVLSGDGGYLFDFFGDKIISYLLISGSAAG 388
>TC218814
Length = 881
Score = 35.0 bits (79), Expect = 0.023
Identities = 22/76 (28%), Positives = 34/76 (43%), Gaps = 7/76 (9%)
Frame = +3
Query: 119 WFDFGHDQVFAYLLLAASAA--GTAMVR-----TLKETDTCTASNAFCVQSDIAVALGYA 171
W F DQ+ Y+ AA+ A G AM+ + C FCVQ A+ GYA
Sbjct: 396 WICFLLDQIAVYMTFAANTAAMGAAMLAITGSDAFQWLKVCDKFTRFCVQIGGALLCGYA 575
Query: 172 GFVFLGFTSLLTGFRV 187
+ + S ++ ++V
Sbjct: 576 ASIIMALISTISAYQV 623
>TC231907
Length = 258
Score = 31.6 bits (70), Expect = 0.25
Identities = 15/63 (23%), Positives = 34/63 (53%)
Frame = +1
Query: 125 DQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTG 184
DQV AYLL++++++ ++E T N F + +A + F L +++++G
Sbjct: 7 DQVVAYLLISSTSSAIPATDEMREVTT----NIFTDSAAAGIAFSFFAFCCLALSAVISG 174
Query: 185 FRV 187
+++
Sbjct: 175 YKL 183
>TC226365 weakly similar to UP|Q944E4 (Q944E4) Cellulose synthase-like protein
OsCslE1, partial (12%)
Length = 1722
Score = 30.0 bits (66), Expect = 0.73
Identities = 19/69 (27%), Positives = 34/69 (48%)
Frame = +3
Query: 66 THGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHD 125
T+G H T+ F+ ++ V+ L+ +V V + +G TVFP+ WF
Sbjct: 1068 TYGFSRMSIIHTFTYCFMTMSSLYAVVFILYGIVPQVC-LLKGITVFPKATDPWF----- 1229
Query: 126 QVFAYLLLA 134
VFA++ ++
Sbjct: 1230 AVFAFVYVS 1256
>TC204827 UP|Q9SLQ6 (Q9SLQ6) Actin isoform B, complete
Length = 1733
Score = 28.9 bits (63), Expect = 1.6
Identities = 15/53 (28%), Positives = 29/53 (54%)
Frame = -2
Query: 93 YSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRT 145
YS F+ + W++ R A + P + ++F G ++LL ASA + ++R+
Sbjct: 1303 YSPFD-IHICWKVLRDARIDPPIQTLYFLSGGATTLIFMLLGASAVISLLIRS 1148
>BF425927
Length = 427
Score = 28.1 bits (61), Expect = 2.8
Identities = 19/64 (29%), Positives = 29/64 (44%), Gaps = 2/64 (3%)
Frame = +1
Query: 11 GADGGDNNNNHRSSSPRFHSTMAE--HKLRRFNSLILVFRLISFSFSLASSVFMLTNTHG 68
GADGGD R S+ AE + ++ + F + + S +L SS F+ +
Sbjct: 22 GADGGDREEQQHGRDDRSRSSWAEVVSQDQQVPFQLPCFLIFASSPNLISSFFIFAGSPS 201
Query: 69 SDSP 72
S SP
Sbjct: 202SSSP 213
>TC222160
Length = 515
Score = 28.1 bits (61), Expect = 2.8
Identities = 16/49 (32%), Positives = 26/49 (52%)
Frame = -2
Query: 115 VLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSD 163
VL+ WFD AY+L++ +AGT + ++ DT T A V ++
Sbjct: 184 VLECWFDSSQ----AYILISFKSAGTLAMLRREQNDTNTKIAATLVTTE 50
>TC203461
Length = 577
Score = 28.1 bits (61), Expect = 2.8
Identities = 16/48 (33%), Positives = 24/48 (49%)
Frame = +1
Query: 152 CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRFHL 199
C++S C S + + LG F+F+ SL F + CF+ G F L
Sbjct: 415 CSSSPFSCSISLLLIVLGICLFLFIN*YSLGFIFVITCFLFLGFMFDL 558
>TC209489
Length = 623
Score = 26.9 bits (58), Expect = 6.2
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = +2
Query: 9 RNGADGGDNNNNHRSSSPRFHSTMAEHKLRRF 40
+N A+ DNNN SP+ S + KLR++
Sbjct: 191 KNQAEADDNNNTDTFLSPKALSELLSTKLRKY 286
>TC215794 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyruvate
translocator, partial (76%)
Length = 1693
Score = 26.9 bits (58), Expect = 6.2
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Frame = +3
Query: 23 SSSPRF---HSTMAEHKLRRFNSLILVFRLISFSFSLAS 58
SSSP++ +S H R+ +L+ FRL FSFS S
Sbjct: 60 SSSPQYKTQNSHNPTHHARQKRALVFRFRLFLFSFSFRS 176
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,707,947
Number of Sequences: 63676
Number of extensions: 129738
Number of successful extensions: 1004
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 989
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 997
length of query: 199
length of database: 12,639,632
effective HSP length: 92
effective length of query: 107
effective length of database: 6,781,440
effective search space: 725614080
effective search space used: 725614080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0180a.7


