

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.6
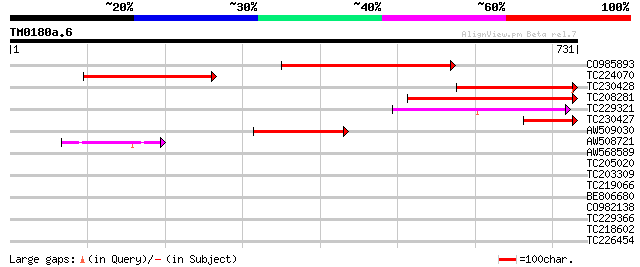
(731 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO985893 343 1e-94
TC224070 similar to UP|Q8RX89 (Q8RX89) AT5g23390/T32G24_2, parti... 273 2e-73
TC230428 weakly similar to UP|Q8RX89 (Q8RX89) AT5g23390/T32G24_2... 258 7e-69
TC208281 221 7e-58
TC229321 143 3e-34
TC230427 123 3e-28
AW509030 105 5e-23
AW508721 65 1e-10
AW568589 32 1.0
TC205020 similar to GB|AAR99874.1|41323411|AY518291 strubbelig r... 31 2.2
TC203309 homologue to UP|O64438 (O64438) ARG10, complete 29 6.5
TC219066 29 6.5
BE806680 29 8.5
CO982138 29 8.5
TC229366 similar to UP|AVR3_CHICK (P56733) Avidin-related protei... 29 8.5
TC218602 similar to GB|AAB66486.1|2318131|AF014824 histone deace... 29 8.5
TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 29 8.5
>CO985893
Length = 829
Score = 343 bits (881), Expect = 1e-94
Identities = 177/225 (78%), Positives = 195/225 (86%)
Frame = -1
Query: 351 KANVRRDYWLDISLEILRVHKFIRKYYLKETQKAEVLARAILGIYRYRAVREAFKYFSSH 410
KAN+RRDYWLDISLEILR HKFIRKYYLKE QK+EVLARA+LGI+RYRAVREAF++FSSH
Sbjct: 826 KANLRRDYWLDISLEILRAHKFIRKYYLKEVQKSEVLARALLGIFRYRAVREAFRFFSSH 647
Query: 411 YKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSKRSSPRNVDTKKQLVVSPASAVALF 470
YKTLL+FNLAE LPRGD+IL+T+S SLTNL S KR P VDTK+Q VSP + +ALF
Sbjct: 646 YKTLLTFNLAETLPRGDIILQTMSKSLTNLAAVSVKRDIPVTVDTKRQPAVSPVAVMALF 467
Query: 471 RLGFKSDKFADIYEETTVVGDIRVGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGID 530
LGFKS K DI EE T V DIRVGEI+ LE+AVKKSL DTGKAEAAQATVDQVKVEGID
Sbjct: 466 YLGFKSKKVTDICEEATFVSDIRVGEIHPLEVAVKKSLLDTGKAEAAQATVDQVKVEGID 287
Query: 531 TNVAVMKELLFPVIESANRLERLASWKDFYKSTAFLLLTSYMILR 575
TNVAVMKELLFPVI SANRL+ LASWKDFYKS AFLLL+ YMI+R
Sbjct: 286 TNVAVMKELLFPVIVSANRLQLLASWKDFYKSAAFLLLSCYMIIR 152
>TC224070 similar to UP|Q8RX89 (Q8RX89) AT5g23390/T32G24_2, partial (16%)
Length = 521
Score = 273 bits (699), Expect = 2e-73
Identities = 136/173 (78%), Positives = 153/173 (87%), Gaps = 2/173 (1%)
Frame = +1
Query: 96 LTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETPSVHT--DETPPPSSK 153
LTYAR+LLEFCSYKALHKL +SD+L D DFRRLT+DMMLAWE PSVHT D SSK
Sbjct: 1 LTYARHLLEFCSYKALHKLIHNSDFLNDNDFRRLTFDMMLAWEAPSVHTLPDNPTSSSSK 180
Query: 154 DEHGGDDDEASLFYSSSTNMAVQVDDQKTVGLEAFSRIAPVCAPIADVITVHNLFDALTS 213
+E GD+D+ASLFYSSSTNMA+QVDD+KTVGLEAFSRIAPVC PIADV+TVHN+F ALTS
Sbjct: 181 EETAGDEDDASLFYSSSTNMALQVDDKKTVGLEAFSRIAPVCIPIADVVTVHNIFHALTS 360
Query: 214 SSGRRLHFLVYDKYIRFLDKVIKNSKNVLASSVGSLQLAEEEIVLDVDGTIPT 266
+S RLHFLVYDKY+RFLDKVIKNSKNV+A+S G+LQLAE EI+LDVDGTIPT
Sbjct: 361 TSAHRLHFLVYDKYLRFLDKVIKNSKNVMATSAGNLQLAEGEIILDVDGTIPT 519
>TC230428 weakly similar to UP|Q8RX89 (Q8RX89) AT5g23390/T32G24_2, partial
(15%)
Length = 748
Score = 258 bits (659), Expect = 7e-69
Identities = 126/161 (78%), Positives = 143/161 (88%), Gaps = 6/161 (3%)
Frame = +3
Query: 577 WIHYLLPSIFVVVAILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLI 636
WI Y +PSIF+ +AILMLWRRH RKGR LEAFIVTPPPNRN VEQLLTLQEAITQFESLI
Sbjct: 6 WIQYFIPSIFMFMAILMLWRRHLRKGRPLEAFIVTPPPNRNAVEQLLTLQEAITQFESLI 185
Query: 637 QAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCR 696
QA NI+LLK+RALLLA+LPQATEKVALLLVF+AAVFA+VPPKYI+ V+F E+YTREMP R
Sbjct: 186 QAANIILLKLRALLLAILPQATEKVALLLVFLAAVFAYVPPKYILLVVFVEFYTREMPYR 365
Query: 697 KESSDRWVRRMREWWIRIPAAPVELIKP------DECKKRK 731
KESSDRW+RR+REWW RIPAAPV+L+KP +E KK+K
Sbjct: 366 KESSDRWIRRIREWWDRIPAAPVQLVKPVHESKKNESKKKK 488
>TC208281
Length = 807
Score = 221 bits (564), Expect = 7e-58
Identities = 115/219 (52%), Positives = 151/219 (68%)
Frame = +1
Query: 513 KAEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYKSTAFLLLTSYM 572
K +AQATVD VKV+GIDTN+AVMKELLFP+ E A L+ LA W D KS F L SY+
Sbjct: 7 KVISAQATVDGVKVDGIDTNLAVMKELLFPLNELAKSLQSLAYWDDPRKSLVFCLFFSYI 186
Query: 573 ILRGWIHYLLPSIFVVVAILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQF 632
I RGW+ Y + + +++A+ M+ R F +GRS+ V PP N +EQLL +Q A++Q
Sbjct: 187 IYRGWLGYAVALVLLLLAVFMIITRCFSQGRSVPEVKVIAPPPLNTMEQLLAVQNAVSQA 366
Query: 633 ESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTRE 692
E LIQ GN++LLK R LLL++ PQATEK+A L+ A + AF+P KYII ++F E +TR
Sbjct: 367 EQLIQDGNVILLKFRGLLLSIFPQATEKLAFGLLSTALILAFLPTKYIILLLFLEIFTRY 546
Query: 693 MPCRKESSDRWVRRMREWWIRIPAAPVELIKPDECKKRK 731
P RK S++R RR++EWW IPAAPV L + E KKRK
Sbjct: 547 SPPRKASTERLTRRLKEWWFSIPAAPVTLERDKEEKKRK 663
>TC229321
Length = 947
Score = 143 bits (360), Expect = 3e-34
Identities = 75/233 (32%), Positives = 134/233 (57%), Gaps = 4/233 (1%)
Frame = +1
Query: 494 VGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERL 553
V +++L+E A K S E QAT+D ++GI +N+ + KEL+FP E+L
Sbjct: 31 VADLSLIERAAKTSKHKYHIVEKTQATIDAATLQGIPSNIDLFKELMFPFTLIVKNFEKL 210
Query: 554 ASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFRK----GRSLEAFI 609
W++ + + AFL LT +I R + Y+ P + +++A+ ML R ++ GRS
Sbjct: 211 RHWEEPHLTIAFLGLTYTIIYRNLLSYMFPMMLMILAVGMLTIRALKEQGRLGRSFGEVT 390
Query: 610 VTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIA 669
+ P N +++++ +++A+ E+ +Q N+ LLK+R++LL+ PQ T +VAL+L+ A
Sbjct: 391 IRDQPPSNTIQKIIAVKDAMRDVENFMQQVNVFLLKIRSILLSGHPQITTEVALVLISSA 570
Query: 670 AVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELI 722
+ +P KYI + F+ +TRE+ R+E ++ +RE W +PA PV ++
Sbjct: 571 TILLIIPFKYIFSFLLFDMFTRELEFRREMVKKFRSFLRERWHTVPAVPVSIL 729
>TC230427
Length = 457
Score = 123 bits (308), Expect = 3e-28
Identities = 56/70 (80%), Positives = 65/70 (92%), Gaps = 1/70 (1%)
Frame = +3
Query: 663 LLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELI 722
LLLVF+AAVFAFVPPKYI+ V+F E+YTREMP RKESSDRW+RR+REWW+RIPAAPV+L+
Sbjct: 3 LLLVFLAAVFAFVPPKYILLVVFVEFYTREMPYRKESSDRWIRRIREWWVRIPAAPVQLV 182
Query: 723 KPD-ECKKRK 731
KPD E KKRK
Sbjct: 183 KPDHESKKRK 212
>AW509030
Length = 443
Score = 105 bits (263), Expect = 5e-23
Identities = 54/124 (43%), Positives = 81/124 (64%), Gaps = 1/124 (0%)
Frame = +2
Query: 315 IKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWLDISLEILRVHKFIR 374
++P TGP GA+L+DKA++Y ST ++E V EFPE ++ RRD+WL + EI+ +H+F+
Sbjct: 2 VEPAATGPWGAQLYDKAIIYDSTDLSETVVLEFPELTSSTRRDHWLALVREIMFLHQFLS 181
Query: 375 KYYLK-ETQKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLAEALPRGDMILETL 433
KY +K Q E+ AR ILGI R +A RE + L F+L + +P+GD +LE L
Sbjct: 182 KYQIKCPIQTWELHARTILGIVRLQAAREMLRISPPVPTKFLIFSLYDEIPKGDYVLEEL 361
Query: 434 SNSL 437
++SL
Sbjct: 362 ADSL 373
>AW508721
Length = 448
Score = 65.1 bits (157), Expect = 1e-10
Identities = 47/143 (32%), Positives = 74/143 (50%), Gaps = 9/143 (6%)
Frame = +3
Query: 68 RSSKILGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSD-YLADKDF 126
R S+ L S E+L F+ E+ Y R L+EFCS K ++++ + + + D F
Sbjct: 6 RCSQKLDTSVEKLVEDFEGGWN---PEMGNYCRKLVEFCSGKGVNEMCHNIEGNINDSSF 176
Query: 127 RRLTYDMMLAWETPSVHTDETPPPSSKDEH--------GGDDDEASLFYSSSTNMAVQVD 178
RLTYDMMLAWE PS + +E +K++ + D+ LFYS M + V+
Sbjct: 177 SRLTYDMMLAWERPSYYDEEPMETVAKEKEERKITLNTTQELDDIPLFYSDI--MPLLVN 350
Query: 179 DQKTVGLEAFSRIAPVCAPIADV 201
++ VG +AF + + +ADV
Sbjct: 351 NEPNVGEDAFVWLGSLVPLVADV 419
>AW568589
Length = 407
Score = 32.0 bits (71), Expect = 1.0
Identities = 20/43 (46%), Positives = 27/43 (62%)
Frame = +1
Query: 250 QLAEEEIVLDVDGTIPTQPVLQHIGISAWPGRLTLTNYALYFE 292
QL EE I+ G+ +PVL+ A PG+LTLT+ A+YFE
Sbjct: 259 QLFEERILCI--GSNSKRPVLKWENNMA*PGKLTLTDKAIYFE 381
>TC205020 similar to GB|AAR99874.1|41323411|AY518291 strubbelig receptor
family 6 {Arabidopsis thaliana;} , partial (50%)
Length = 1841
Score = 30.8 bits (68), Expect = 2.2
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = +2
Query: 87 ELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETPSVH 143
++ LG+ L Y L E CS +HK KS++ L D DF D LA P+ +
Sbjct: 890 KIALGIARALEY---LHEVCSPSVVHKNIKSANILLDTDFNPHLSDSGLASYIPNAN 1051
>TC203309 homologue to UP|O64438 (O64438) ARG10, complete
Length = 1052
Score = 29.3 bits (64), Expect = 6.5
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = +3
Query: 679 YIIFVIFFEYYTREMPCRKESSDRWVRRMREWW 711
Y++F +F RE C S+ +W RR R WW
Sbjct: 87 YLLF--YFL*RERERKCWGSSAVQWCRRRRSWW 179
>TC219066
Length = 586
Score = 29.3 bits (64), Expect = 6.5
Identities = 17/63 (26%), Positives = 28/63 (43%), Gaps = 4/63 (6%)
Frame = +2
Query: 554 ASWKDFYKSTAFLLLTSYMILRGWIHYLLPS----IFVVVAILMLWRRHFRKGRSLEAFI 609
A W D + L+ S ++L G + + +V+ IL +W + KGR AF+
Sbjct: 191 AFWPDVHFRIVMALIVSQIVLMGLLTTKKAASSTPFLIVLPILTIWFHRYCKGRFESAFV 370
Query: 610 VTP 612
P
Sbjct: 371 KCP 379
>BE806680
Length = 383
Score = 28.9 bits (63), Expect = 8.5
Identities = 23/83 (27%), Positives = 42/83 (49%), Gaps = 4/83 (4%)
Frame = +1
Query: 231 LDKVIKNSKNVLASSV-GSLQLAEEEIVLDVDGTIPTQPVLQHIG-ISAWPGRLTLTNYA 288
+DK I++ NVL+ S+ GSL +I+ T + +L ++ P + +L+N A
Sbjct: 52 IDKAIEDGVNVLSMSIGGSLMEYYRDIIAIGSFTAMSHGILVSTSTVNGGPSQGSLSNVA 231
Query: 289 LYFESLGVGVHEK--PVRYDLGT 309
+ ++G G ++ P LGT
Sbjct: 232 PWITTVGAGTIDRDCPAYITLGT 300
>CO982138
Length = 807
Score = 28.9 bits (63), Expect = 8.5
Identities = 25/93 (26%), Positives = 41/93 (43%), Gaps = 6/93 (6%)
Frame = -2
Query: 626 QEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIF 685
+EAI ++ N L K+RA+ P T K+A+LL +A + + + F
Sbjct: 632 EEAIWVLRLILPYLNEFLSKLRAMFSGD-PGTTIKLAVLLFVLARCGSSITIWKMAKFGF 456
Query: 686 FEYYTREMPCRKESSDR------WVRRMREWWI 712
F +T C S+ W+RR R+ W+
Sbjct: 455 FGVFTGPKICSSYSAQLTAFANFWIRRFRDAWV 357
>TC229366 similar to UP|AVR3_CHICK (P56733) Avidin-related protein 3 precursor,
partial (13%)
Length = 1728
Score = 28.9 bits (63), Expect = 8.5
Identities = 12/41 (29%), Positives = 21/41 (50%)
Frame = -3
Query: 664 LLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWV 704
+ +F+ +F + IF++FF Y+ +K SS WV
Sbjct: 1318 IYLFLYFIFIYFKANTFIFILFFTYFFSSY*LKK*SSSLWV 1196
>TC218602 similar to GB|AAB66486.1|2318131|AF014824 histone deacetylase
{Arabidopsis thaliana;} , partial (21%)
Length = 771
Score = 28.9 bits (63), Expect = 8.5
Identities = 17/64 (26%), Positives = 30/64 (46%)
Frame = +3
Query: 115 SKSSDYLADKDFRRLTYDMMLAWETPSVHTDETPPPSSKDEHGGDDDEASLFYSSSTNMA 174
+K+S +L ++ +L ++ PSV E PP S E D D+ + ++M
Sbjct: 90 NKNSRHLLEEIRSKLLENLSKLQHAPSVQFQERPPDSDLGEADEDHDDGDEPWDPDSDMD 269
Query: 175 VQVD 178
V V+
Sbjct: 270 VDVE 281
>TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (71%)
Length = 1889
Score = 28.9 bits (63), Expect = 8.5
Identities = 15/41 (36%), Positives = 24/41 (57%)
Frame = -1
Query: 408 SSHYKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSKRS 448
S+H+ L +L+ G +L T+S+SLT T +SS R+
Sbjct: 614 STHFNPLTRASLSSMDKTGGPLLATISSSLTRPTTSSSPRA 492
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,794,414
Number of Sequences: 63676
Number of extensions: 405524
Number of successful extensions: 2479
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 2441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2473
length of query: 731
length of database: 12,639,632
effective HSP length: 104
effective length of query: 627
effective length of database: 6,017,328
effective search space: 3772864656
effective search space used: 3772864656
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0180a.6


