

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.4
(342 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
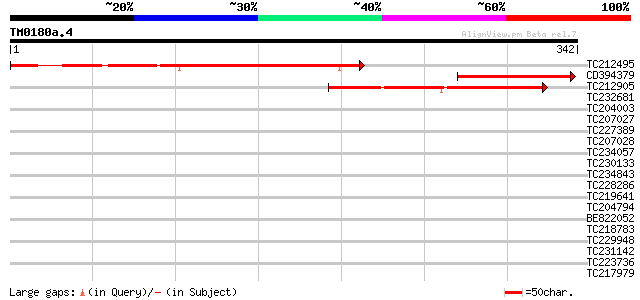
Score E
Sequences producing significant alignments: (bits) Value
TC212495 weakly similar to UP|AF9_HUMAN (P42568) AF-9 protein, p... 239 1e-63
CD394379 115 4e-26
TC212905 similar to GB|AAC03244.1|2896826|DADIASHB1 hunchback pr... 102 3e-22
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 39 0.003
TC204003 37 0.013
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 37 0.017
TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 35 0.049
TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%) 35 0.049
TC234057 similar to UP|Q9LSV7 (Q9LSV7) Gb|AAD21483.1, partial (3%) 33 0.19
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 32 0.42
TC234843 similar to UP|WSC2_YEAST (P53832) Cell wall integrity a... 32 0.54
TC228286 weakly similar to GB|AAH20129.1|18044890|BC020129 Robo4... 32 0.54
TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 32 0.54
TC204794 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 31 0.71
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 31 0.71
TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%) 31 0.71
TC229948 similar to UP|Q9MAC6 (Q9MAC6) T4P13.15 protein, partial... 31 0.93
TC231142 weakly similar to UP|Q7PC53 (Q7PC53) Chitinase B, parti... 31 0.93
TC223736 weakly similar to UP|Q8VYJ8 (Q8VYJ8) AT4g27310/M4I22_12... 30 1.2
TC217979 weakly similar to UP|Q6NLH4 (Q6NLH4) At5g54470, partial... 30 1.6
>TC212495 weakly similar to UP|AF9_HUMAN (P42568) AF-9 protein, partial (7%)
Length = 703
Score = 239 bits (611), Expect = 1e-63
Identities = 126/218 (57%), Positives = 156/218 (70%), Gaps = 4/218 (1%)
Frame = +2
Query: 1 MVVPHSMTLPSSSLSSPIGVKERNFNMPTSYCCLHKDKPRKQFLNLSLSSNLSLLVGDSK 60
M +PHS+ LP SS+ SP CCL K KP L L+ + + +
Sbjct: 104 MALPHSIILPFSSIISP--------------CCLPKHKPTNFTLPFKLNGDSCRSI---R 232
Query: 61 APRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPIN--DPYLMTLEERQEWRRKI 118
P RVQALKSDG K +EASSD+D D DD++DAP N DPYLM+ EER EWRR I
Sbjct: 233 IPSRVQALKSDGGKWKKRGQEASSDTDDD-DDDDDAPQRFNKNDPYLMSPEERLEWRRNI 409
Query: 119 RQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKIT 178
RQV+D KP+V+EE DP EKKKK+EKL++DYPLVVDE+DP+WPEDADGWGFSLGQFF+KIT
Sbjct: 410 RQVLDRKPDVEEELDPLEKKKKLEKLLEDYPLVVDEDDPDWPEDADGWGFSLGQFFDKIT 589
Query: 179 IKDNKKAKDDDDDDDEGNK--VVWQDDDYIRPIKDIKS 214
IK+ KK DDDDD+D+ ++ ++WQDD+YIRPIKDIK+
Sbjct: 590 IKNKKKDDDDDDDNDDVDRPEIMWQDDNYIRPIKDIKT 703
>CD394379
Length = 693
Score = 115 bits (287), Expect = 4e-26
Identities = 56/71 (78%), Positives = 66/71 (92%)
Frame = -2
Query: 271 AVVETELVAALKVSVFPEMIFTKSGKILFRDKAIRNADELSKIMAYFYFGAAKPPCLNSF 330
AVVETELVAAL+VS+FPE+IFTK+GKILFRDKAIR+A+E SK+MAYFY+GAAKP CLNS
Sbjct: 692 AVVETELVAALQVSIFPEIIFTKAGKILFRDKAIRSAEEWSKVMAYFYYGAAKPSCLNSL 513
Query: 331 TDFQEDIPSLV 341
T QE+IPS+V
Sbjct: 512 TYSQENIPSIV 480
>TC212905 similar to GB|AAC03244.1|2896826|DADIASHB1 hunchback protein
{Drosophila adiastola;} , partial (16%)
Length = 644
Score = 102 bits (254), Expect = 3e-22
Identities = 52/134 (38%), Positives = 84/134 (61%), Gaps = 2/134 (1%)
Frame = +2
Query: 193 DEGNKVVWQDDDYIRPIKDIKSSEWEETVFKDISPLIILVHNRYRRPKENERIRDELEKA 252
++G ++ W+ + +++I EWE F SPLI+LV RY R +N +I ELEKA
Sbjct: 50 EDGYEIDWEGEIDDNWVQEINCLEWESFAFHP-SPLIVLVFERYNRASDNWKILKELEKA 226
Query: 253 VHIIWNC--RLPSPRCVALDAVVETELVAALKVSVFPEMIFTKSGKILFRDKAIRNADEL 310
+ + W RLP PR V +D +E +L ALKV P+++F + ++++R+K +R ADEL
Sbjct: 227 IKVYWRAKDRLP-PRAVKIDINIERDLAYALKVRECPQILFLRGHRVVYREKELRTADEL 403
Query: 311 SKIMAYFYFGAAKP 324
+++A+FY+ A KP
Sbjct: 404 VQMIAFFYYNAKKP 445
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 38.9 bits (89), Expect = 0.003
Identities = 44/151 (29%), Positives = 67/151 (44%), Gaps = 19/151 (12%)
Frame = +1
Query: 79 KREASSDSDSDS-----DDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESD 133
+R S D D D+ DDE+DAP +D EE + +R D + E+ D
Sbjct: 193 RRRLSFDDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDED 372
Query: 134 PEEKKKKMEK--LMKDY---PLVVDEE---------DPNWPEDADGWGFSLGQFFNKITI 179
E+++++ E+ L +Y PLV EE + N E+ + G+ ++
Sbjct: 373 EEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDDEDGE-KSEAPP 549
Query: 180 KDNKKAKDDDDDDDEGNKVVWQDDDYIRPIK 210
K + KDD DDDD G +DD RP K
Sbjct: 550 KRKRSDKDDSDDDDGG------EDDDERPSK 624
>TC204003
Length = 809
Score = 37.0 bits (84), Expect = 0.013
Identities = 15/44 (34%), Positives = 26/44 (59%)
Frame = +2
Query: 62 PRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYL 105
P++V+A K +G ++ GK+E + D++ PPP+ DP L
Sbjct: 155 PKKVEA-KKEGEKKEEGKKEEGKKEEKKEDEKKKDPPPVPDPVL 283
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 36.6 bits (83), Expect = 0.017
Identities = 41/173 (23%), Positives = 77/173 (43%), Gaps = 9/173 (5%)
Frame = +3
Query: 57 GDSKAPRRVQALKSDGA---HRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQE 113
GD +V+ ++DG +K K++ D D +D E + EE++E
Sbjct: 72 GDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKDGKTDAEKIKKKEDDGK---DDEEKKE 242
Query: 114 WRRKIRQVMDMKPNVQEESDPEE-KKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQ 172
++K ++ K +E+ +E K+K +K D D E+ +D + +
Sbjct: 243 KKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKGDG----DGEEKKEKKDKEKEKKKEKK 410
Query: 173 FFNKITIKDNKKAKDDDDDDDEGNKVVWQD-----DDYIRPIKDIKSSEWEET 220
++ T +K K+D+ +DDEGNK +D D+ + KD + E E++
Sbjct: 411 DKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKEKDHKKEKKDKEEGEKEDS 569
>TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (63%)
Length = 1144
Score = 35.0 bits (79), Expect = 0.049
Identities = 31/118 (26%), Positives = 57/118 (48%), Gaps = 2/118 (1%)
Frame = +3
Query: 83 SSDSD--SDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKK 140
+SD D +DSD++++ P IN T + E + + +V + K V + S ++ K
Sbjct: 360 NSDEDEFTDSDEDDEDVPLIN-----TENGKPETKAEDLKVPESKKAVAKASGSAKQVKV 524
Query: 141 MEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKV 198
+E P +EED + D D +G S + + + D++ +DDD++ KV
Sbjct: 525 VE------PKKDNEEDSDDDSDDDDFGSSDEEMEDADSDSDDESGDEDDDEETPPKKV 680
>TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%)
Length = 490
Score = 35.0 bits (79), Expect = 0.049
Identities = 35/118 (29%), Positives = 52/118 (43%), Gaps = 8/118 (6%)
Frame = +1
Query: 111 RQEWRRKIRQVMDMKPNVQEES---DPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWG 167
R+ +RK R M + + +E EEKK+K EK K DEE
Sbjct: 70 RRRRKRKRRTKMKKQTH*REREKMVSGEEKKEKKEKEKKKEKKDKDEETD---------- 219
Query: 168 FSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQD-----DDYIRPIKDIKSSEWEET 220
T+K+ K K+D+ +DDEGNK +D D+ + KDI+ E E++
Sbjct: 220 ----------TLKE--KGKNDEGEDDEGNKKKKKDKKEKEKDHKKEKKDIEEGEKEDS 357
>TC234057 similar to UP|Q9LSV7 (Q9LSV7) Gb|AAD21483.1, partial (3%)
Length = 493
Score = 33.1 bits (74), Expect = 0.19
Identities = 34/139 (24%), Positives = 59/139 (41%), Gaps = 6/139 (4%)
Frame = +3
Query: 75 RKSGKREASSDSDSDSDDENDAPPPINDPYLMTL-----EERQEWRRKIRQVMDMKPNVQ 129
+ SG+ S S S+ +N D LM + E+ + ++ D ++
Sbjct: 6 QNSGRESIESGSISNFQLQNS----YKDRELMNVNIGSTEDETDMNSELDSDSDKLSDIF 173
Query: 130 EESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDD 189
E E K EK PL +DE D N+PE +DG + ++++K KDD+
Sbjct: 174 ETDSDTENFVKEEK-----PLYLDEFD-NFPEQSDGEMNDFEEHMRQMSLKSKNMEKDDN 335
Query: 190 -DDDDEGNKVVWQDDDYIR 207
DE +K+ Q +++
Sbjct: 336 LPKLDEVDKIFLQATSFLK 392
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 32.0 bits (71), Expect = 0.42
Identities = 17/52 (32%), Positives = 25/52 (47%)
Frame = +3
Query: 176 KITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIKSSEWEETVFKDISP 227
K T ++NK A D +DDDD+ N +DD+ +D + E D P
Sbjct: 246 KHTSEENKDASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDP 401
>TC234843 similar to UP|WSC2_YEAST (P53832) Cell wall integrity and stress
response component 2 precursor, partial (4%)
Length = 407
Score = 31.6 bits (70), Expect = 0.54
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = -1
Query: 176 KITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKD 211
KIT K + DDDDDDD+ + W+D+ +R + +
Sbjct: 158 KIT-KSSYMLVDDDDDDDDDGQASWEDEKGLRHVNN 54
>TC228286 weakly similar to GB|AAH20129.1|18044890|BC020129 Robo4 protein
{Mus musculus;} , partial (6%)
Length = 920
Score = 31.6 bits (70), Expect = 0.54
Identities = 27/98 (27%), Positives = 34/98 (34%), Gaps = 16/98 (16%)
Frame = +2
Query: 114 WRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDY--------PLVVDEEDPNWPEDADG 165
W+ V PN P + + KMEKL K Y L V + +W
Sbjct: 119 WQEVADAVSSRCPNASPPKTPVQCRHKMEKLRKRYRTEIQRARSLPVSRFNSSWAH---- 286
Query: 166 WGFSLGQFFNK--------ITIKDNKKAKDDDDDDDEG 195
F L K I D+ +DDDDDD G
Sbjct: 287 --FKLMDSMEKGPSPVKPDINDSDHNNTDNDDDDDDAG 394
>TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 942
Score = 31.6 bits (70), Expect = 0.54
Identities = 30/112 (26%), Positives = 48/112 (42%)
Frame = +1
Query: 83 SSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKME 142
+SDSD D D+E D +D EE E D +EE+DPE+ +
Sbjct: 376 ASDSDEDGDEEEDD----DDADDQDDEEGDE---------DFSGEGEEEADPEDDPEANG 516
Query: 143 KLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDE 194
D D++D +D D G G+ D ++ +D+D++D+E
Sbjct: 517 AGGSDSD--GDDDDDGGDDDDDDNGDDDGE--------DEEEGEDEDEEDEE 642
>TC204794 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1037
Score = 31.2 bits (69), Expect = 0.71
Identities = 23/71 (32%), Positives = 35/71 (48%)
Frame = +1
Query: 81 EASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKK 140
++ DSD D DDE+D + + + E +E RK RQ Q+E EE K K
Sbjct: 463 KSDDDSDDDEDDEDDTEALLAELEQIKKERAEEKLRKERQ--------QQE---EELKVK 609
Query: 141 MEKLMKDYPLV 151
+L++ PL+
Sbjct: 610 EAELLRGNPLL 642
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 31.2 bits (69), Expect = 0.71
Identities = 38/139 (27%), Positives = 55/139 (39%)
Frame = -1
Query: 83 SSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKME 142
+SDSD D D+E+D +D EE E D +EE+DP
Sbjct: 527 ASDSDEDEDEEDD-----HDTNDQNDEEGDE---------DFSGEGEEEADP-------- 414
Query: 143 KLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQD 202
E+DP +A+G G S D+ + +DDDDD++G+ D
Sbjct: 413 -----------EDDP----EANGAGGS----------NDDDDSDEDDDDDNDGD-----D 324
Query: 203 DDYIRPIKDIKSSEWEETV 221
D K+ + E EE V
Sbjct: 323 DGEDEDEKEGEDEEDEEEV 267
>TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%)
Length = 870
Score = 31.2 bits (69), Expect = 0.71
Identities = 36/145 (24%), Positives = 53/145 (35%), Gaps = 16/145 (11%)
Frame = +3
Query: 76 KSGKREASSDSDSDSD---DENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEES 132
K GK ASS S SD DE++AP P + + + D + + S
Sbjct: 180 KKGKPAASSSSSESSDDNSDEDEAPKTKVAPAAGKNGHASTKKTQPSESSDSDSSDSDSS 359
Query: 133 DPEEKKKKMEKLMKDYPLVV--------DEEDPNWPEDADGWGFSLGQFFNKITIKDNKK 184
E K KK K L V +++ + D D +K + + KK
Sbjct: 360 SDEGKSKKKPTTAKLPTLPVAPAKKVESSDDESSESSDEDNDAKPAVTAVSKPSARAQKK 539
Query: 185 AKDDDDDD-----DEGNKVVWQDDD 204
+ D DD D+G + DDD
Sbjct: 540 VESSDSDDSSSDEDKGKMDIDDDDD 614
>TC229948 similar to UP|Q9MAC6 (Q9MAC6) T4P13.15 protein, partial (16%)
Length = 1798
Score = 30.8 bits (68), Expect = 0.93
Identities = 34/156 (21%), Positives = 59/156 (37%), Gaps = 5/156 (3%)
Frame = +1
Query: 55 LVGDSKAPRRVQALK---SDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEER 111
L D P R + L +D + +E + +S+SDD D + P M ++
Sbjct: 10 LTWDDDEPLRAKTLNRKLTDEQLAEFEVKEFLTSDESESDDSKDNNETDDQPDKMA--KK 183
Query: 112 QEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLG 171
+ R + Q D E + + + ++D + E+ E W L
Sbjct: 184 RAKYRALLQSGDDSDGGNEHDNAQNMEVTFNTGLEDLSKYIMEKKDKKSETV--WDAYLS 357
Query: 172 QFFNKITIKDNKK--AKDDDDDDDEGNKVVWQDDDY 205
+ K NK+ + DDDD DD + + DD+
Sbjct: 358 KKSEKKKAGKNKRKYSSDDDDSDDTDQEATEEADDF 465
>TC231142 weakly similar to UP|Q7PC53 (Q7PC53) Chitinase B, partial (3%)
Length = 1130
Score = 30.8 bits (68), Expect = 0.93
Identities = 36/127 (28%), Positives = 52/127 (40%), Gaps = 7/127 (5%)
Frame = +1
Query: 74 HRKSGKREASSDSDSDSDDENDAP-PPINDPYLMTLEERQEWRRKIR-QVMDMKPNVQE- 130
H S RE+ D D DDE +P PI+ +E Q M+ NVQ
Sbjct: 235 HSSSIGRESLRSMDEDWDDEMASPVTPIHMGAANNSNSIEEAESDDNVQAMESDDNVQAM 414
Query: 131 ESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADG----WGFSLGQFFNKITIKDNKKAK 186
ESD E+ + + D V+ + NW +G W L + D+ +A
Sbjct: 415 ESDDNEQAAESD----DNGQAVEYDQGNWEIGFNGAPSRWSRDLVGMSD-----DDVEAS 567
Query: 187 DDDDDDD 193
+DDDD+D
Sbjct: 568 EDDDDND 588
>TC223736 weakly similar to UP|Q8VYJ8 (Q8VYJ8) AT4g27310/M4I22_120, partial
(38%)
Length = 807
Score = 30.4 bits (67), Expect = 1.2
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = +3
Query: 181 DNKKAKDDDDDDDEGNKVV 199
D A+DDDDDDDE N+VV
Sbjct: 471 DGGVAEDDDDDDDEENQVV 527
>TC217979 weakly similar to UP|Q6NLH4 (Q6NLH4) At5g54470, partial (33%)
Length = 461
Score = 30.0 bits (66), Expect = 1.6
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Frame = +2
Query: 181 DNKKAKDDDDDDDEG-NKVV-WQDDDYIRPIKDIKSSEWEE 219
D +DDD+DD++G N+VV W P SEW E
Sbjct: 335 DEPDTEDDDNDDEDGDNQVVPWSSSPPPPPPSSSSGSEWSE 457
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,014,875
Number of Sequences: 63676
Number of extensions: 231095
Number of successful extensions: 2022
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 1553
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1806
length of query: 342
length of database: 12,639,632
effective HSP length: 98
effective length of query: 244
effective length of database: 6,399,384
effective search space: 1561449696
effective search space used: 1561449696
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0180a.4


