

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0178.14
(599 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
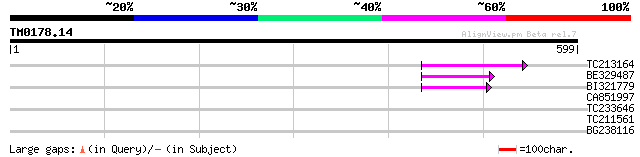
TC213164 68 1e-11
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 60 4e-09
BI321779 48 1e-05
CA851997 37 0.033
TC233646 32 1.0
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 29 5.2
BG238116 weakly similar to SP|Q9M7Q7|PCN1_ Proliferating cellula... 28 8.9
>TC213164
Length = 446
Score = 67.8 bits (164), Expect = 1e-11
Identities = 37/112 (33%), Positives = 57/112 (50%)
Frame = +3
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDGLN F K WD + F++ G + N + LIPKV +P S+
Sbjct: 78 KSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKRSNAFFLALIPKVHDP*SLNE 257
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQE 547
Y+PIS YK++++++ R K L +I Q+ F+ + N++IA E
Sbjct: 258 YKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEGRHTLHNVVIANE 413
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 59.7 bits (143), Expect = 4e-09
Identities = 27/77 (35%), Positives = 42/77 (54%)
Frame = -2
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDGLN F K+ W+ + F + G P N + + LIPKV +P+++
Sbjct: 342 KSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALIPKVKHPQALND 163
Query: 496 YRPISCCNFTYKVISRI 512
+RPIS YK++++I
Sbjct: 162 FRPISLIGCVYKIVAKI 112
>BI321779
Length = 421
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/74 (33%), Positives = 33/74 (43%)
Frame = +2
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDG + L K W+ + E + F LP N + + LI K NP I
Sbjct: 143 KSPGPDGYSLLLIKIFWEVIKEEVINFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQ 322
Query: 496 YRPISCCNFTYKVI 509
Y PIS YK++
Sbjct: 323 YCPISLVGCLYKIV 364
>CA851997
Length = 636
Score = 36.6 bits (83), Expect = 0.033
Identities = 23/86 (26%), Positives = 39/86 (44%), Gaps = 2/86 (2%)
Frame = +1
Query: 440 PDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRPI 499
PDG N FY W E +A + + G PS +N+T + +I ++
Sbjct: 316 PDGFNLGFYHRFWGMCGEDVFQACCMWLAEGAFPSSVNDTTIAIILEI**S*RYERS*TN 495
Query: 500 SCCNFTYKV--ISRIIVTRLKGGLNQ 523
CN +K+ +S ++ RLK L++
Sbjct: 496 LLCNGGFKILFLSEVLAKRLKNVLDK 573
>TC233646
Length = 978
Score = 31.6 bits (70), Expect = 1.0
Identities = 26/66 (39%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Frame = -3
Query: 472 LPSKINETVVTLIPKVPNPESITHYRPISCCNFTYKVISRII-VTRLKGGLNQLITLNQS 530
LPS I+ + LIPK+ N E + Y IS CN T +++SR T K + I NQ
Sbjct: 844 LPSIIS---LVLIPKIDNLEILK*Y*SISLCNVTTRLLSRSS*PTSSKSFPS*PIARNQC 674
Query: 531 AFIGVK 536
+FI K
Sbjct: 673 SFIPSK 656
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 29.3 bits (64), Expect = 5.2
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +1
Query: 557 RASKEHVALKLDMSKAYERVEWSFL 581
R++K + K+D KAY+ V W+FL
Sbjct: 46 RSNKSCLVFKVDYEKAYDSVSWNFL 120
>BG238116 weakly similar to SP|Q9M7Q7|PCN1_ Proliferating cellular nuclear
antigen 1 (PCNA 1). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (22%)
Length = 544
Score = 28.5 bits (62), Expect = 8.9
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Frame = +2
Query: 468 STGHLPSKINETVVTLIPKVPNPESITHYR--PISCCNFTYKVISRII 513
STG +N T+ L+P + PE YR PI+ T +RI+
Sbjct: 92 STGFSLKALNSTLCALVPLLLRPEGFNPYRWNPITSWGLTLNTWARIL 235
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.350 0.155 0.548
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,228,173
Number of Sequences: 63676
Number of extensions: 411811
Number of successful extensions: 3623
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 2645
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 945
Number of HSP's gapped (non-prelim): 2790
length of query: 599
length of database: 12,639,632
effective HSP length: 103
effective length of query: 496
effective length of database: 6,081,004
effective search space: 3016177984
effective search space used: 3016177984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0178.14


