

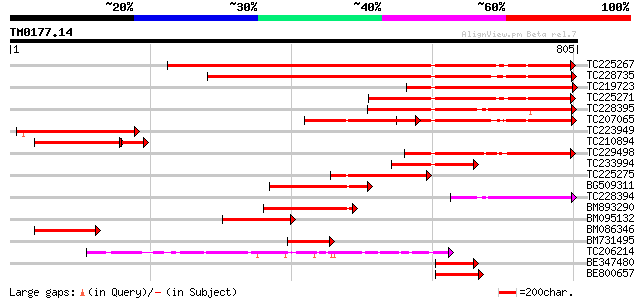
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.14
(805 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 547 e-156
TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (... 469 e-132
TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-li... 382 e-106
TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment... 260 1e-69
TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_3... 256 2e-68
TC207065 beta-glucosidase 226 4e-59
TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 201 1e-51
TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%) 159 2e-49
TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, parti... 187 1e-47
TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partia... 165 6e-41
TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylos... 147 2e-35
BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 132 8e-31
TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 131 1e-30
BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 130 3e-30
BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum v... 127 1e-29
BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabido... 117 2e-26
BM731495 weakly similar to PIR|T49928|T499 beta-glucosidase-like... 115 6e-26
TC206214 beta-D-glucan exohydrolase [Glycine max] 98 2e-20
BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus pe... 83 4e-16
BE800657 weakly similar to GP|4100433|gb|A beta-glucosidase {Gly... 66 5e-11
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 547 bits (1409), Expect = e-156
Identities = 278/584 (47%), Positives = 382/584 (64%), Gaps = 4/584 (0%)
Frame = +3
Query: 224 GDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMC 283
G+ L V+ACCKH+TAYDL+ W R++FNA VS+QDLEDTY PF+ CV +G+ + +MC
Sbjct: 18 GNHLKVAACCKHYTAYDLDNWNGVDRFHFNAKVSKQDLEDTYDVPFKACVLEGQVASVMC 197
Query: 284 SYNEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLK 342
SYN+VNG P CA DLL R W GYI SDCD+V F+ Q Y K+ E+A AE +K
Sbjct: 198 SYNQVNGKPTCADPDLLRNTIRGQWRLNGYIVSDCDSVGVFFDNQHYTKTPEEAAAEAIK 377
Query: 343 AGTDINCGTYMLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGP 402
AG D++CG ++ HT SA+ +G + E D++ AL NL SVQ+RLG+FDG+P T YG LGP
Sbjct: 378 AGLDLDCGPFLAIHTDSAIRKGLISENDLNLALANLISVQMRLGMFDGEPSTQPYGNLGP 557
Query: 403 HDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGI 461
DVCTS H+ LALEAAR+ IVLL+N LPL+ + ++ V+GP A T + G Y+G+
Sbjct: 558 RDVCTSAHQQLALEAARESIVLLQNKGNSLPLSPSRLRTIGVVGPNADATVTMIGNYAGV 737
Query: 462 PCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLET 521
C + +G+A Y K ++ GC ++C + F A ARQAD +V+V G+D T+E
Sbjct: 738 ACGYTTPLQGIARYV-KTAHQVGCKGVACRGNELFGAAETIARQADAIVLVMGLDQTVEA 914
Query: 522 EDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPG 581
E DRV LLLPG Q +LV+ +A A+K PVIL++ GGP+D+SFA+ + I +ILWVGYPG
Sbjct: 915 ETRDRVGLLLPGLQQELVTRVARAAKGPVILLIMSGGPVDISFAKNDPKISAILWVGYPG 1094
Query: 582 EAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESF-TNVPMNDMSMRADPSRGYPGRTYR 640
+AGG A+A++IFG +NP GRLPMTWYP+ + VPM +M MR +P+ GYPGRTYR
Sbjct: 1095QAGGTAIADVIFGTTNP-----GGRLPMTWYPQGYLAKVPMTNMDMRPNPTTGYPGRTYR 1259
Query: 641 FYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKS-LSDQAEKEVYGVDYVQ 699
FY G V+ FGHGLSYS FS+ AP +VS+ ++ +L S LS +A K V +
Sbjct: 1260FYKGPVVFPFGHGLSYSRFSHSLALAPKQVSVPIMSLQALTNSTLSSKAVK----VSHAN 1427
Query: 700 VDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSK 759
D+ SL H+ V N G +DG+H +++FS+ P + S QLVGF + H ++
Sbjct: 1428CDD-----SLEMEFHVDVKNEGSMDGTHTLLIFSQ-PPHGKWSQIKQLVGFHKTHVLAGS 1589
Query: 760 SIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
+ V C+HLS D+ G R P G H L +GDV+H++S++
Sbjct: 1590KQRVKVGVHVCKHLSVVDQFGVRRIPTGEHELHIGDVKHSISVQ 1721
>TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (65%)
Length = 1599
Score = 469 bits (1206), Expect = e-132
Identities = 256/527 (48%), Positives = 326/527 (61%), Gaps = 3/527 (0%)
Frame = +1
Query: 281 LMCSYNEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAE 339
+MCSYN+VNG P CA DLL GV R W GYI SDCD+V +++ Q Y K+ E+A A
Sbjct: 10 VMCSYNKVNGKPTCADPDLLKGVVRGEWKLNGYIVSDCDSVEVLYKDQHYTKTPEEAAAI 189
Query: 340 VLKAGTDINCGTYMLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGK 399
+ AG D+NCG ++ ++T AV+QG + E I+ A+ N F+ +RLG FDGDPR YG
Sbjct: 190 SILAGLDLNCGRFLGQYTEGAVKQGLIDEASINNAVTNNFATLMRLGFFDGDPRKQPYGN 369
Query: 400 LGPHDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGY 458
LGP DVCT E++ LA EAARQGIVLLKN LPLN SLAVIGP A T + G Y
Sbjct: 370 LGPKDVCTQENQELAREAARQGIVLLKNSPASLPLNAKAIKSLAVIGPNANATRVMIGNY 549
Query: 459 SGIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTT 518
GIPC S +GL +A SYA+GC D+ C + +A + A AD V+V G
Sbjct: 550 EGIPCKYISPLQGLTAFA-PTSYAAGCLDVRC-PNPVLDDAKKIAASADATVMVVGASLA 723
Query: 519 LETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVG 578
+E E DRV++LLPG+Q LVS +A ASK PVILV+ GG +DVSFA+ N I SILWVG
Sbjct: 724 IEAESLDRVNILLPGQQQLLVSEVANASKGPVILVIMSGGGMDVSFAKNNNKITSILWVG 903
Query: 579 YPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTN-VPMNDMSMRADPSRGYPGR 637
YPGEAGG A+A++IFG NP +GRLPMTWYP+S+ + VPM +M+MR DP+ GYPGR
Sbjct: 904 YPGEAGGAAIADVIFGFHNP-----SGRLPMTWYPQSYVDKVPMTNMNMRPDPATGYPGR 1068
Query: 638 TYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDY 697
TYRFY G V+ FG GLSYS +K + AP VS+ R S E +D
Sbjct: 1069TYRFYKGETVFAFGDGLSYSSIVHKLVKAPQLVSVQLAEDHVCRSS-------ECKSIDV 1227
Query: 698 VQVDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVS 757
V C +L F +H+ + N G + +H V LFS P V +PQ L+GF +VH +
Sbjct: 1228VGE----HCQNLVFDIHLRIKNKGKMSSAHTVFLFST-PPAVHNAPQKHLLGFEKVHLIG 1392
Query: 758 SKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
S VD C+ LS DE G R LG H+L VGD++H +S+ I
Sbjct: 1393KSEALVSFKVDVCKDLSIVDELGNRKVALGQHLLHVGDLKHPLSVMI 1533
>TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-like protein
(AT5g10560/F12B17_90), partial (19%)
Length = 1013
Score = 382 bits (982), Expect = e-106
Identities = 188/242 (77%), Positives = 206/242 (84%)
Frame = +1
Query: 564 FAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMND 623
FAE+N I SI+W+GYPGEAGGKALAEIIFGE NP AGRLPMTWYPE+FTNVPMN+
Sbjct: 1 FAEKNPQIASIIWLGYPGEAGGKALAEIIFGEFNP-----AGRLPMTWYPEAFTNVPMNE 165
Query: 624 MSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKS 683
MSMRADPSRGYPGRTYRFYTG RVYGFGHGLS+S FSY FLSAPSK+SLSR K RK
Sbjct: 166 MSMRADPSRGYPGRTYRFYTGGRVYGFGHGLSFSDFSYNFLSAPSKISLSRTIKDGSRKR 345
Query: 684 LSDQAEKEVYGVDYVQVDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSP 743
L Q E EVYGVDYV V++L +CN LSF VHISV NLG LDGSHVVMLFSK PKVV+GSP
Sbjct: 346 LLYQVENEVYGVDYVPVNQLQNCNKLSFSVHISVMNLGGLDGSHVVMLFSKGPKVVDGSP 525
Query: 744 QTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
+TQLVGFSR+HT+SSK ETSILV PCEHLSFAD+QGKR+ PLG H LSVGD+EH VSIE
Sbjct: 526 ETQLVGFSRLHTISSKPTETSILVHPCEHLSFADKQGKRILPLGPHTLSVGDLEHVVSIE 705
Query: 804 IF 805
I+
Sbjct: 706 IY 711
>TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment), partial
(55%)
Length = 952
Score = 260 bits (665), Expect = 1e-69
Identities = 138/296 (46%), Positives = 188/296 (62%), Gaps = 2/296 (0%)
Frame = +3
Query: 510 VIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQ 569
V+V G+D T+E E DRV LLLPG Q +LV+ +A A+K PVILV+ GGP+DVSFA+ N
Sbjct: 9 VLVMGLDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKNNP 188
Query: 570 LIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESF-TNVPMNDMSMRA 628
I +ILWVGYPG+AGG A+A++IFG +NP GRLPMTWYP+ + VPM +M MR
Sbjct: 189 KISAILWVGYPGQAGGTAIADVIFGATNP-----GGRLPMTWYPQGYLAKVPMTNMDMRP 353
Query: 629 DPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKS-LSDQ 687
+P+ GYPGRTYRFY G V+ FGHGLSYS FS AP +VS+ ++ +L S LS +
Sbjct: 354 NPATGYPGRTYRFYKGPVVFPFGHGLSYSRFSQSLALAPKQVSVQILSLQALTNSTLSSK 533
Query: 688 AEKEVYGVDYVQVDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQL 747
A K V + D+ SL H+ V N G +DG+H +++FSK P + S QL
Sbjct: 534 AVK----VSHANCDD-----SLETEFHVDVKNEGSMDGTHTLLIFSK-PPPGKWSQIKQL 683
Query: 748 VGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
V F + H + + V C+HLS D+ G R P G H L +GD++H+++++
Sbjct: 684 VTFHKTHVPAGSKQRLKVNVHSCKHLSVVDQFGVRRIPTGEHELHIGDLKHSINVQ 851
>TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial
(22%)
Length = 1011
Score = 256 bits (655), Expect = 2e-68
Identities = 136/301 (45%), Positives = 189/301 (62%), Gaps = 5/301 (1%)
Frame = +3
Query: 509 VVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERN 568
VV+V G+D + E E HDR L LPGKQ +L+ S+A ASK PV+LVL GGP+D++ A+ +
Sbjct: 18 VVLVMGLDQSQERESHDREYLGLPGKQEELIKSVARASKRPVVLVLLCGGPVDITSAKFD 197
Query: 569 QLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRA 628
+ ILW GYPGE GG ALA+++FG+ NP G+LP+TWYP+ F VPM DM MRA
Sbjct: 198 DKVGGILWAGYPGELGGVALAQVVFGDHNP-----GGKLPITWYPKDFIKVPMTDMRMRA 362
Query: 629 DPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKSLSDQA 688
DP+ GYPGRTYRFYTG +VY FG+GLSY+ +SYK L SLS T + +S +
Sbjct: 363 DPASGYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLL------SLSHSTL-HINQSSTHLM 521
Query: 689 EKEVYGVDYVQVDELL--SCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKV---VEGSP 743
+ + Y V EL +C ++ + + VTN G+L G H V+LF + KV G+P
Sbjct: 522 TQNSETIRYKLVSELAEETCQTMLLSIALGVTNRGNLAGKHPVLLFVRQGKVRNINNGNP 701
Query: 744 QTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
QLVGF V + ++++ + PCEHLS A+E G V G+++ VGD E+ + +
Sbjct: 702 VKQLVGFQSVKVNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLFIVGDQEYPIEVT 881
Query: 804 I 804
+
Sbjct: 882 V 884
>TC207065 beta-glucosidase
Length = 1328
Score = 226 bits (575), Expect = 4e-59
Identities = 120/256 (46%), Positives = 158/256 (60%), Gaps = 1/256 (0%)
Frame = +2
Query: 550 VILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPM 609
VILV+ GG +DVSFA+ N I SILWVGYPGEAGG A+A++IFG NP +GRLPM
Sbjct: 488 VILVIMSGGGMDVSFAKSNDKITSILWVGYPGEAGGAAIADVIFGFYNP-----SGRLPM 652
Query: 610 TWYPESFTN-VPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPS 668
TWYP+++ N VPM +M+MRADP+ GYPGRTYRFY G V+ FG G+S+S +K + AP
Sbjct: 653 TWYPQAYVNKVPMTNMNMRADPATGYPGRTYRFYKGETVFSFGDGISFSSIEHKIVKAPQ 832
Query: 669 KVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSCNSLSFPVHISVTNLGDLDGSHV 728
VS+ R S E +D DE C +L+F +H+ V N G + SHV
Sbjct: 833 LVSVPLAEDHECRSS-------ECMSLDI--ADE--HCQNLAFDIHLGVKNTGKMSTSHV 979
Query: 729 VMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGN 788
V+LF P V +PQ L+GF +VH + VD C+ LS DE G R PLG
Sbjct: 980 VLLFFT-PPDVHNAPQKHLLGFEKVHLPGKSEAQVRFKVDVCKDLSVVDELGNRKVPLGQ 1156
Query: 789 HVLSVGDVEHTVSIEI 804
H+L VG+++H +S+ +
Sbjct: 1157HLLHVGNLKHPLSLRV 1204
Score = 143 bits (360), Expect = 3e-34
Identities = 81/166 (48%), Positives = 104/166 (61%), Gaps = 1/166 (0%)
Frame = +1
Query: 419 RQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAK 477
RQGIVLLKN LPLN SLAVIGP A T + G Y GIPC+ S + L
Sbjct: 4 RQGIVLLKNSPGSLPLNAKTIKSLAVIGPNANATRVMIGNYEGIPCNYISPLQTLTALVP 183
Query: 478 KISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMD 537
SYA+GC ++ C ++ +A + A AD VI+ G +E E DR+++LLPG+Q
Sbjct: 184 T-SYAAGCPNVQC-ANAELDDATQIAASADATVIIVGASLAIEAESLDRINILLPGQQQL 357
Query: 538 LVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEA 583
LVS +A ASK PVILV+ GG +DVSFA+ N I SILW+GYPG++
Sbjct: 358 LVSEVANASKGPVILVIMSGGGMDVSFAKSNDKITSILWIGYPGDS 495
>TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (20%)
Length = 638
Score = 201 bits (511), Expect = 1e-51
Identities = 99/180 (55%), Positives = 130/180 (72%), Gaps = 5/180 (2%)
Frame = +3
Query: 10 FIFFVVLI---HFTTSIEDFACKRPHHSRY-KFCDTSLSIPTRAHSLVSLLTLPEKIQQL 65
FI F++L H ++ ++C +S Y FC+T L I RA LVS LTL EK+ QL
Sbjct: 24 FISFLLLTLHHHAESTQPPYSCDSSSNSPYYPFCNTRLPITKRAQDLVSRLTLDEKLAQL 203
Query: 66 SNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWF 125
N A +IPRLGIP+YQWWSE+LHG+A G G+ F+G + +AT FPQVI++AASF+ LW+
Sbjct: 204 VNTAPAIPRLGIPSYQWWSEALHGVADAGFGIRFNGTIKSATSFPQVILTAASFDPNLWY 383
Query: 126 LIASAVAVEARAMFNVGQA-GLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVR 184
I+ + EARA++N GQA G+TFWAPN+NVFRDPRWGRGQET GEDP++ + Y V YV+
Sbjct: 384 QISKTIGKEARAVYNAGQATGMTFWAPNINVFRDPRWGRGQETAGEDPLMNAKYGVAYVK 563
>TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%)
Length = 738
Score = 159 bits (401), Expect(2) = 2e-49
Identities = 74/127 (58%), Positives = 95/127 (74%)
Frame = +3
Query: 36 YKFCDTSLSIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGP 95
+KFC+T + I R L++ LTLPEKI+ + NNA ++PRLGI Y+WWSE+LHG++ GP
Sbjct: 174 FKFCNTHVPIHVRVQDLIARLTLPEKIRLVVNNAIAVPRLGIQGYEWWSEALHGVSNVGP 353
Query: 96 GVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVAVEARAMFNVGQAGLTFWAPNVNV 155
G F GA AT FPQVI +AASFN++LW I V+ EARAM+N GQAGLT+W+PNVN+
Sbjct: 354 GTKFGGAFPGATMFPQVISTAASFNQSLWQEIGRVVSDEARAMYNGGQAGLTYWSPNVNI 533
Query: 156 FRDPRWG 162
FRDP G
Sbjct: 534 FRDPPVG 554
Score = 56.6 bits (135), Expect(2) = 2e-49
Identities = 25/38 (65%), Positives = 29/38 (75%)
Frame = +1
Query: 160 RWGRGQETPGEDPMVASAYAVDYVRGLQGAGGVKNVFG 197
RWGRGQETPGEDP +A+ YA YV+GLQG G K+ G
Sbjct: 547 RWGRGQETPGEDPTLAAKYAXSYVKGLQGDGARKSPQG 660
>TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial (24%)
Length = 792
Score = 187 bits (476), Expect = 1e-47
Identities = 98/245 (40%), Positives = 150/245 (61%), Gaps = 2/245 (0%)
Frame = +1
Query: 561 DVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESF-TNV 619
D++FA+ N I ILW GYPG+AGG A+A+I+FG +NP G+LP+TWYPE + T +
Sbjct: 1 DITFAKNNPRIVGILWAGYPGQAGGAAIADILFGTANP-----GGKLPVTWYPEEYLTKL 165
Query: 620 PMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGS 679
PM +M+MRA S GYPGRTYRFY G VY FGHGL+Y+ F + SAP+ VS+ G
Sbjct: 166 PMTNMAMRATKSAGYPGRTYRFYNGPVVYPFGHGLTYTHFVHTLASAPTVVSVP--LNGH 339
Query: 680 LRKSLSDQAEKEVYGVDYVQVDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPK-V 738
R ++++ + + + V + + C+ LS + + + N+G DG+H +++FS P
Sbjct: 340 RRANVTNISNRAI-RVTHAR------CDKLSISLEVDIKNVGSRDGTHTLLVFSAPPAGF 498
Query: 739 VEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEH 798
+ + QLV F ++H + + + C+ LS D+ G R PLG H ++GDV+H
Sbjct: 499 GHWALEKQLVAFEKIHVPAKGLQRVGVNIHVCKLLSVVDKSGIRRIPLGEHSFNIGDVKH 678
Query: 799 TVSIE 803
+VS++
Sbjct: 679 SVSLQ 693
>TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (15%)
Length = 462
Score = 165 bits (418), Expect = 6e-41
Identities = 79/124 (63%), Positives = 92/124 (73%)
Frame = +2
Query: 542 IAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGF 601
+A ASK PVILVL GGPLD++ A+ N I ILW GYPGE GG ALA+IIFG+ NP
Sbjct: 2 VAEASKKPVILVLLSGGPLDITSAKYNHKIGGILWAGYPGELGGIALAQIIFGDHNP--- 172
Query: 602 NAAGRLPMTWYPESFTNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSY 661
GRLP TWYP+ + VPM DM MRADPS GYPGRTYRFY G +VY FG+GLSYS +SY
Sbjct: 173 --GGRLPTTWYPKDYIKVPMTDMRMRADPSTGYPGRTYRFYKGPKVYEFGYGLSYSKYSY 346
Query: 662 KFLS 665
+ +S
Sbjct: 347 ELVS 358
>TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (32%)
Length = 933
Score = 147 bits (371), Expect = 2e-35
Identities = 71/143 (49%), Positives = 97/143 (67%)
Frame = +3
Query: 456 GGYSGIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGI 515
G Y+G+ C + +G+A Y K ++ GC ++C + F A ARQ D V+V G+
Sbjct: 18 GNYAGVACGYTTPLQGIARYVKT-AHQVGCRGVACRGNELFGAAEIIARQVDATVLVMGL 194
Query: 516 DTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSIL 575
D T+E E DRV LLLPG Q +LV+ +A A+K PVILV+ GGP+DVSFA+ N I +IL
Sbjct: 195 DQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKNNPKISAIL 374
Query: 576 WVGYPGEAGGKALAEIIFGESNP 598
WVGYPG+AGG A+A++IFG +NP
Sbjct: 375 WVGYPGQAGGTAIADVIFGATNP 443
>BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (31%)
Length = 443
Score = 132 bits (331), Expect = 8e-31
Identities = 68/147 (46%), Positives = 95/147 (64%), Gaps = 1/147 (0%)
Frame = +1
Query: 370 DIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVLLKNDK 429
D++ AL+N +VQ+RLG+FDG+P YG LGP DVC H+ LALEAARQGIVLLKN
Sbjct: 4 DVNGALVNTLTVQMRLGMFDGEPTAHPYGHLGPKDVCKPAHQELALEAARQGIVLLKNTG 183
Query: 430 KFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKISYASGCSDI 488
LPL+ ++AVIGP + T + G Y+G+ C + +G+ YA+ + + GC ++
Sbjct: 184 PVLPLSSQLHRTVAVIGPNSKATITMIGNYAGVACGYTNPLQGIGRYARTV-HQLGCQNV 360
Query: 489 SCNSDGGFAEAIETARQADFVVIVAGI 515
+C +D F AI ARQAD V+V G+
Sbjct: 361 ACKNDKLFGPAINAARQADATVLVMGL 441
>TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (14%)
Length = 659
Score = 131 bits (329), Expect = 1e-30
Identities = 75/182 (41%), Positives = 108/182 (59%), Gaps = 3/182 (1%)
Frame = +2
Query: 626 MRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKSLS 685
MRADP+ GYPGRTYRFYTG +VY FG+GLSY+ +SYK L SLS T + +S +
Sbjct: 2 MRADPASGYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLL------SLSHNTL-HINQSST 160
Query: 686 DQAEKEVYGVDYVQVDELL--SCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKV-VEGS 742
+ + Y V EL +C ++ + + VTN G++ G H V+LF + KV G+
Sbjct: 161 HLTTQNSETIRYKLVSELAEETCQTMLLSIALGVTNHGNMAGKHPVLLFVRQGKVRNNGN 340
Query: 743 PQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSI 802
P QLVGF V + ++++ + PCEHLS A+E G V G+++L VGD E+ + I
Sbjct: 341 PVKQLVGFQSVKLNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLLLVGDQEYPIEI 520
Query: 803 EI 804
+
Sbjct: 521 TV 526
>BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (27%)
Length = 403
Score = 130 bits (326), Expect = 3e-30
Identities = 66/134 (49%), Positives = 90/134 (66%), Gaps = 1/134 (0%)
Frame = +1
Query: 361 VEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQ 420
V++G + E D++ ALLN +VQ+RLG++DG+P + Y LGP DVCT H+ LALEAARQ
Sbjct: 1 VKKGLISEADVNGALLNTLTVQMRLGMYDGEPSSHPYNNLGPRDVCTQSHQELALEAARQ 180
Query: 421 GIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKI 479
GIVLLKN LPL+ G ++AVIGP + VT + G Y+GI C S +G+ Y K I
Sbjct: 181 GIVLLKNKGPSLPLSTRRGRTVAVIGPNSNVTFTMIGNYAGIACGYTSPLQGIGTYTKTI 360
Query: 480 SYASGCSDISCNSD 493
Y GC++++C D
Sbjct: 361 -YEHGCANVACTDD 399
>BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum vulgare},
partial (2%)
Length = 428
Score = 127 bits (320), Expect = 1e-29
Identities = 58/104 (55%), Positives = 76/104 (72%)
Frame = +1
Query: 303 ARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVE 362
AR W F GYITSDC AV+ + E QGY K+AEDA+A+V +AG D+ CG Y+ +H SAV
Sbjct: 10 ARQQWKFDGYITSDCGAVSIIHEKQGYAKTAEDAIADVFRAGMDVECGDYITKHAKSAVF 189
Query: 363 QGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVC 406
Q K+ IDRAL NLFS+++RLGLFDG+P +G +GP++VC
Sbjct: 190 QKKLPISQIDRALQNLFSIRIRLGLFDGNPTKLPFGTIGPNEVC 321
>BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabidopsis
thaliana}, partial (11%)
Length = 427
Score = 117 bits (294), Expect = 2e-26
Identities = 55/93 (59%), Positives = 70/93 (75%)
Frame = -3
Query: 36 YKFCDTSLSIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGP 95
Y FC+T L I RA LVS LTL EK+ QL N A +IPRLGIP+YQWWSE+LHG+A G
Sbjct: 374 YPFCNTRLPITKRAQDLVSRLTLDEKLAQLVNTAPAIPRLGIPSYQWWSEALHGVADAGF 195
Query: 96 GVSFDGAVSAATDFPQVIVSAASFNRTLWFLIA 128
G+ F+G + +AT FPQVI++AASF+ LW+ I+
Sbjct: 194 GIRFNGTIKSATSFPQVILTAASFDPNLWYQIS 96
>BM731495 weakly similar to PIR|T49928|T499 beta-glucosidase-like protein -
Arabidopsis thaliana, partial (13%)
Length = 421
Score = 115 bits (289), Expect = 6e-26
Identities = 54/67 (80%), Positives = 62/67 (91%)
Frame = +2
Query: 395 GKYGKLGPHDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMAVTNKL 454
G++GKLGP DVCT EHKTLAL+AARQGIVLLKNDKKFLPL+R+ G+SLAVIGP+A T KL
Sbjct: 5 GRFGKLGPKDVCTQEHKTLALDAARQGIVLLKNDKKFLPLDRDIGASLAVIGPLATTTKL 184
Query: 455 GGGYSGI 461
GGGYSG+
Sbjct: 185 GGGYSGM 205
>TC206214 beta-D-glucan exohydrolase [Glycine max]
Length = 1647
Score = 97.8 bits (242), Expect = 2e-20
Identities = 137/546 (25%), Positives = 236/546 (43%), Gaps = 24/546 (4%)
Frame = +1
Query: 109 FPQVIVSAASFNRTLWFLIASAVAVEARAMFNVGQAGLTF-WAPNVNVFRDPRWGRGQET 167
FP + + + L I A A+E RA G+ + +AP + V RDPRWGR E+
Sbjct: 4 FPHNVGLGVTRDPVLIKKIGEATALEVRA------TGIPYVFAPCIAVCRDPRWGRCYES 165
Query: 168 PGEDPMVASAYAVDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNG-SFDSDSDDGDDGDS 226
EDP + + + GLQG D G S G F + +
Sbjct: 166 YSEDPKIVKTMT-EIIPGLQG---------------DIPGNSIKGVPFVAGKNK------ 279
Query: 227 LMVSACCKHFTAYDLEKWGQFARYNFN-AVVSQQDLEDTYQPPFRGCVQQGKASCLMCSY 285
V+AC KH+ L G N N ++S L + P + + +G S +M SY
Sbjct: 280 --VAACAKHY----LGDGGTNKGINENNTLISYNGLLSIHMPAYYDSIIKG-VSTVMISY 438
Query: 286 NEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAVATVFE--YQGYVKSAEDAVAEVLK 342
+ NG+ A++ L+ G +N FKG++ SD + + + Y S + V+
Sbjct: 439 SSWNGMKMHANKKLITGYLKNKLHFKGFVISDWQGIDRITSPPHANYSYSVQAGVS---- 606
Query: 343 AGTDINCG----TYMLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDG---DPRTG 395
AG D+ T + V+ + ID A+ + V+ +GLF+ DP
Sbjct: 607 AGIDMIMVPFNYTEFIDELTRQVKNNIIPISRIDDAVARILRVKFVMGLFENPYADPSLA 786
Query: 396 KYGKLGPHDVCTSEHKTLALEAARQGIVLLKNDKKF----LPLNRNYGSSLAVIGPMAVT 451
+LG + EH+ +A EA R+ +VLLKN K + LPL + + + V G A
Sbjct: 787 N--QLG-----SKEHREIAREAVRKSLVLLKNGKSYKKPLLPLPKK-STKILVAGSHA-- 936
Query: 452 NKLG---GGYS----GIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETAR 504
N LG GG++ G+ + + + + K+ + + N D F ++
Sbjct: 937 NNLGYQCGGWTITWQGLGGNDLTSGTTILDAVKQTVDPATEVVFNENPDKNFVKSY---- 1104
Query: 505 QADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSF 564
+ D+ ++V G T ET D ++L + ++++ A + V+LV TG + +
Sbjct: 1105KFDYAIVVVGEHTYAETFG-DSLNLTMADPGPSTITNVCGAIRCVVVLV-TGRPVVIKPY 1278
Query: 565 AERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDM 624
+ I +++ PG G+ +A++++G+ + G+L TW+ ++ +PMN
Sbjct: 1279LPK---IDALVAAWLPG-TEGQGVADVLYGD-----YEFTGKLARTWF-KTVDQLPMNVG 1428
Query: 625 SMRADP 630
DP
Sbjct: 1429DKHYDP 1446
>BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus persica},
partial (17%)
Length = 473
Score = 83.2 bits (204), Expect = 4e-16
Identities = 37/62 (59%), Positives = 47/62 (75%), Gaps = 1/62 (1%)
Frame = +3
Query: 605 GRLPMTWYPESFT-NVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKF 663
G+LPMTWYP+ + N+PM +M+MRA S+GYPGRTYRFY G VY FG+GLSY+ F +
Sbjct: 288 GKLPMTWYPQGYIKNLPMTNMAMRASRSKGYPGRTYRFYNGPVVYPFGYGLSYTHFVHTL 467
Query: 664 LS 665
S
Sbjct: 468 TS 473
Score = 33.9 bits (76), Expect = 0.29
Identities = 13/19 (68%), Positives = 17/19 (89%)
Frame = +2
Query: 580 PGEAGGKALAEIIFGESNP 598
PG+AGG A+A+I+FG SNP
Sbjct: 2 PGQAGGAAIADILFGTSNP 58
>BE800657 weakly similar to GP|4100433|gb|A beta-glucosidase {Glycine max},
partial (19%)
Length = 220
Score = 66.2 bits (160), Expect = 5e-11
Identities = 32/69 (46%), Positives = 43/69 (61%), Gaps = 1/69 (1%)
Frame = +2
Query: 605 GRLPMTWYPESF-TNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKF 663
GRLPMTWYP+ + VP+ ++ M +PS YP +TYRFY ++ F H LSYS FS
Sbjct: 8 GRLPMTWYPQRYLAKVPITNIDMHPNPSTRYP*KTYRFYK*PVIFPFKHKLSYSRFSQIL 187
Query: 664 LSAPSKVSL 672
P +VS+
Sbjct: 188 SLTPKQVSI 214
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,284,048
Number of Sequences: 63676
Number of extensions: 494222
Number of successful extensions: 2557
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 2474
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2508
length of query: 805
length of database: 12,639,632
effective HSP length: 105
effective length of query: 700
effective length of database: 5,953,652
effective search space: 4167556400
effective search space used: 4167556400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0177.14


