

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
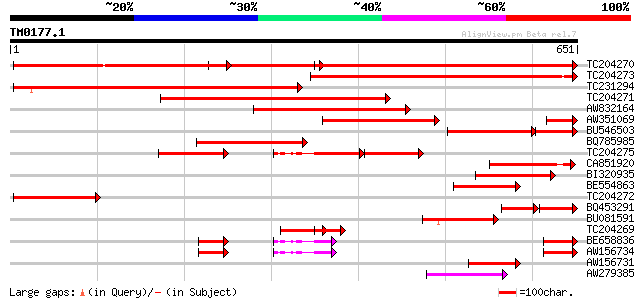
Query= TM0177.1
(651 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204270 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carbo... 587 e-168
TC204273 similar to UP|Q8W2C2 (Q8W2C2) Phosphoenolpyruvate carbo... 556 e-159
TC231294 similar to UP|Q8W505 (Q8W505) Phosphoenolpyruvate carbo... 534 e-152
TC204271 homologue to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate car... 525 e-149
AW832164 316 2e-86
AW351069 homologue to GP|13785467|db phosphoenolpyruvate carboxy... 269 3e-72
BU546503 183 2e-67
BQ785985 241 1e-63
TC204275 homologue to UP|PPCK_CUCSA (P42066) Phosphoenolpyruvate... 154 3e-61
CA851920 similar to GP|13785471|dbj phosphoenolpyruvate carboxyk... 150 1e-36
BI320935 similar to GP|13785471|dbj phosphoenolpyruvate carboxyk... 147 1e-35
BE554863 145 6e-35
TC204272 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carbo... 143 3e-34
BQ453291 81 4e-33
BU081591 homologue to SP|Q9SLZ0|PPCK_ Phosphoenolpyruvate carbox... 132 4e-31
TC204269 homologue to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate car... 87 2e-17
BE658836 homologue to GP|13785467|dbj phosphoenolpyruvate carbox... 77 2e-14
AW156734 homologue to PIR|T05900|T059 phosphoenolpyruvate carbox... 77 2e-14
AW156731 weakly similar to GP|13785467|dbj phosphoenolpyruvate c... 73 5e-13
AW279385 69 5e-12
>TC204270 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase ,
partial (94%)
Length = 2872
Score = 587 bits (1514), Expect = e-168
Identities = 282/301 (93%), Positives = 292/301 (96%)
Frame = +2
Query: 351 GTGKTTLSTDHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVL 410
GTGKTTLSTDHNRYLIGDDEHCW D GVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVL
Sbjct: 1769 GTGKTTLSTDHNRYLIGDDEHCWGDNGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVL 1948
Query: 411 ENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVILLACDAFGVLPP 470
ENVVFDEH REVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHP NVILLACDAFGVLPP
Sbjct: 1949 ENVVFDEHDREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPKNVILLACDAFGVLPP 2128
Query: 471 VSKLNLAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMLHPTKYAAMLAEKME 530
VSKLNLAQTMYHFISGYTALVAGTE+GIKEPQATFSACFGAAFIMLHPTKYAAMLAEKM+
Sbjct: 2129 VSKLNLAQTMYHFISGYTALVAGTEEGIKEPQATFSACFGAAFIMLHPTKYAAMLAEKMQ 2308
Query: 531 QHGATGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLEIPTEV 590
+HGATGWLVNTGWS GSYG+GSRIKLQYTRKIIDAIHSGSLL AE++KT IFGLEIPTEV
Sbjct: 2309 KHGATGWLVNTGWSAGSYGNGSRIKLQYTRKIIDAIHSGSLLNAEYKKTEIFGLEIPTEV 2488
Query: 591 EGVPSEILDPVNTWSDKDAYQETLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPN 650
EGVPSEILDP+NTWS+K AY+ETLLKLAGLFKNNFETFTNYKIGKG+NNLTEEILAAGP
Sbjct: 2489 EGVPSEILDPMNTWSNKKAYKETLLKLAGLFKNNFETFTNYKIGKGNNNLTEEILAAGPI 2668
Query: 651 F 651
F
Sbjct: 2669 F 2671
Score = 399 bits (1026), Expect = e-111
Identities = 202/255 (79%), Positives = 224/255 (87%), Gaps = 5/255 (1%)
Frame = +3
Query: 5 NGNGIATNGLPSIHTQK--NGICHDDSVPTVKANTIDELHSLQKKKSAPGTPISGTQTPF 62
NGNG A NGL I T K NGICHDDS PTVKA TIDELHSLQKKKSAP TP++G Q PF
Sbjct: 204 NGNGTARNGLAKIQTHKKHNGICHDDSGPTVKAQTIDELHSLQKKKSAPTTPVTGFQAPF 383
Query: 63 TS--DPERQQQQLQSISASLASLTRETGPKVVKGDPAKKLENPKTIHQVSHHHIA-PTIA 119
+ + ER +QQLQSISASLASLTRETGP +VKGDPA+K + K H V HHH++ PTIA
Sbjct: 384 ATLTEEERHKQQLQSISASLASLTRETGPNLVKGDPARKSDTQKVTH-VHHHHVSTPTIA 560
Query: 120 VSDSALKFTHVLYNLSPAELYEQAIRYEKGSFVTSTGALATLSGAKTGRSPRDKRVVRDA 179
VSDSALKFTHVLYNLSPAELYEQAI+YEKGSF++STGALATLSGAKTGRSPRDKRVV+D
Sbjct: 561 VSDSALKFTHVLYNLSPAELYEQAIKYEKGSFISSTGALATLSGAKTGRSPRDKRVVKDD 740
Query: 180 VTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVFVNDQFLNWDPENRIKVRIVA 239
+T++DLWWGKGSPNIEMDE +F+VNRERAVDYLNSLDKVFVNDQFLNWD ENRIKVRIV+
Sbjct: 741 LTKNDLWWGKGSPNIEMDEHSFMVNRERAVDYLNSLDKVFVNDQFLNWDTENRIKVRIVS 920
Query: 240 ARAYHSLFMHNMCIR 254
ARAYHSLFMHNM ++
Sbjct: 921 ARAYHSLFMHNM*VK 965
Score = 207 bits (528), Expect = 9e-54
Identities = 103/133 (77%), Positives = 113/133 (84%)
Frame = +2
Query: 229 PENRIKVRIVAARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNRFTHYMTSS 288
P + VR+V + ++ + CIRPTPEELENFGTPDFTIYNAGQFPCNR+THYMTSS
Sbjct: 1166 PPKIVHVRLV*CTKVDN-WVGDRCIRPTPEELENFGTPDFTIYNAGQFPCNRYTHYMTSS 1342
Query: 289 TSVDLNLARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFG 348
TS+DLNLARREMVILGTQYAGEMKKGLF VMHYLMPKR+ILSLHSGCNMGKDGDVALFFG
Sbjct: 1343 TSIDLNLARREMVILGTQYAGEMKKGLFSVMHYLMPKRQILSLHSGCNMGKDGDVALFFG 1522
Query: 349 LSGTGKTTLSTDH 361
LSG +S DH
Sbjct: 1523 LSG----IVSADH 1549
>TC204273 similar to UP|Q8W2C2 (Q8W2C2) Phosphoenolpyruvate carboxykinase
(Fragment), partial (89%)
Length = 1130
Score = 556 bits (1434), Expect = e-159
Identities = 265/306 (86%), Positives = 286/306 (92%)
Frame = +3
Query: 346 FFGLSGTGKTTLSTDHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIR 405
FFGLSGTGKTTLSTDHNRYLIGDDEHCWS+ GV+NIEGGCYAKC+DLS++KEPDIWNAI+
Sbjct: 9 FFGLSGTGKTTLSTDHNRYLIGDDEHCWSENGVANIEGGCYAKCVDLSKDKEPDIWNAIK 188
Query: 406 FGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVILLACDAF 465
FGTVLENVVFDEHTREVD+SDKSVTENTRAAYPIEYIPNAK+PCV PH NVILLACDAF
Sbjct: 189 FGTVLENVVFDEHTREVDFSDKSVTENTRAAYPIEYIPNAKIPCVAPHAKNVILLACDAF 368
Query: 466 GVLPPVSKLNLAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMLHPTKYAAML 525
GVLPPVSKL+LAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIM+HPTKYAAML
Sbjct: 369 GVLPPVSKLSLAQTMYHFISGYTALVAGTEDGIKEPQATFSACFGAAFIMMHPTKYAAML 548
Query: 526 AEKMEQHGATGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLE 585
AEKM++HGATGWLVNTGWSGGSYG G+RIKL YTRKIIDAIHSGSLL A++ KT +FGLE
Sbjct: 549 AEKMQKHGATGWLVNTGWSGGSYGLGNRIKLAYTRKIIDAIHSGSLLNAQYTKTEVFGLE 728
Query: 586 IPTEVEGVPSEILDPVNTWSDKDAYQETLLKLAGLFKNNFETFTNYKIGKGDNNLTEEIL 645
IPT +EGVPSEILDPVNTWSDK AYQ+T LKLA LFK NF+ FT YKIG GD LTEEI+
Sbjct: 729 IPTALEGVPSEILDPVNTWSDKKAYQDTRLKLASLFKKNFDGFTTYKIG-GDQKLTEEIV 905
Query: 646 AAGPNF 651
+AGP F
Sbjct: 906 SAGPIF 923
>TC231294 similar to UP|Q8W505 (Q8W505) Phosphoenolpyruvate carboxykinase,
partial (43%)
Length = 1123
Score = 534 bits (1376), Expect = e-152
Identities = 266/342 (77%), Positives = 290/342 (84%), Gaps = 10/342 (2%)
Frame = +1
Query: 5 NGNGIATNGLPSIHTQKNG------ICHDDSVPTVKANTIDELHSLQKKKSAPGTPISGT 58
N + A GLP I T +CHDDS P V+A TI ELHSLQKKKS P TP+S T
Sbjct: 97 NTSNNARRGLPMIQTNMKPTDSEFQVCHDDSTPPVRAQTIHELHSLQKKKSTPSTPLSAT 276
Query: 59 QTPFT--SDPERQQQQLQSISASLASLTRETGPKVVKGDPAKKLENPKTIHQ--VSHHHI 114
Q PF SD ERQ+QQLQSISASLASLTRE+GPKVVKGDPA K E +H H+
Sbjct: 277 QGPFATVSDEERQKQQLQSISASLASLTRESGPKVVKGDPATKAEKGTHVHHHPQPHYFA 456
Query: 115 APTIAVSDSALKFTHVLYNLSPAELYEQAIRYEKGSFVTSTGALATLSGAKTGRSPRDKR 174
+ VSDSALKFTHVLYNLSPAELYEQAI++EKGSF+T+TGALATLSGAKTGRSPRDKR
Sbjct: 457 STAFDVSDSALKFTHVLYNLSPAELYEQAIKHEKGSFITATGALATLSGAKTGRSPRDKR 636
Query: 175 VVRDAVTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVFVNDQFLNWDPENRIK 234
VVRD TE DLWWGKGSPNIEMDE TFL+NRERAVDYLNSL+KV+VNDQFLNWDPE+RIK
Sbjct: 637 VVRDDTTEHDLWWGKGSPNIEMDEHTFLINRERAVDYLNSLEKVYVNDQFLNWDPEHRIK 816
Query: 235 VRIVAARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNRFTHYMTSSTSVDLN 294
VRIV+ARAYHSLFMHNMCIRPTPEELE FGTPDFTIYNAGQFPCNR+THYMTSSTS+D+N
Sbjct: 817 VRIVSARAYHSLFMHNMCIRPTPEELEEFGTPDFTIYNAGQFPCNRYTHYMTSSTSIDIN 996
Query: 295 LARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCN 336
+AR+EMVILGTQYAGEMKKGLFG+MHYLMPKR ILSLHSGCN
Sbjct: 997 IARKEMVILGTQYAGEMKKGLFGLMHYLMPKRXILSLHSGCN 1122
>TC204271 homologue to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase
, partial (40%)
Length = 801
Score = 525 bits (1353), Expect = e-149
Identities = 247/264 (93%), Positives = 257/264 (96%)
Frame = +3
Query: 174 RVVRDAVTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVFVNDQFLNWDPENRI 233
RVV+D +T++DLWWGKGSPNIEMDE +F+VNRERAVDYLNSLDKVFVNDQFLNWD ENRI
Sbjct: 9 RVVKDDLTKNDLWWGKGSPNIEMDEHSFMVNRERAVDYLNSLDKVFVNDQFLNWDTENRI 188
Query: 234 KVRIVAARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNRFTHYMTSSTSVDL 293
KVRIV+ARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNR+THYMTSSTS+DL
Sbjct: 189 KVRIVSARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAGQFPCNRYTHYMTSSTSIDL 368
Query: 294 NLARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTG 353
NLARREMVILGTQYAGEMKKGLF VMHYLMPKR+ILSLHSGCNMGKDGDVALFFGLSGTG
Sbjct: 369 NLARREMVILGTQYAGEMKKGLFSVMHYLMPKRQILSLHSGCNMGKDGDVALFFGLSGTG 548
Query: 354 KTTLSTDHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENV 413
KTTLSTDHNRYLIGDDEHCW D GVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENV
Sbjct: 549 KTTLSTDHNRYLIGDDEHCWGDNGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENV 728
Query: 414 VFDEHTREVDYSDKSVTENTRAAY 437
VFDEH REVDYSDKSVTENTRAAY
Sbjct: 729 VFDEHDREVDYSDKSVTENTRAAY 800
>AW832164
Length = 544
Score = 316 bits (809), Expect = 2e-86
Identities = 152/181 (83%), Positives = 161/181 (87%)
Frame = +2
Query: 280 RFTHYMTSSTSVDLNLARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGK 339
R+THYMTSST++DLNLARREMVILGTQYAGEMKKGLF M YLMPKR+ILSLHSGCN+G+
Sbjct: 2 RYTHYMTSSTNIDLNLARREMVILGTQYAGEMKKGLFSAMPYLMPKRQILSLHSGCNIGQ 181
Query: 340 DGDVALFFGLSGTGKTTLSTDHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPD 399
DGDVAL+FGLS TGKTTLSTDHNRY IG DEH W D GVS IE GCYAKCIDLSREKEPD
Sbjct: 182 DGDVALYFGLSCTGKTTLSTDHNRY*IGHDEH*WGDNGVSYIECGCYAKCIDLSREKEPD 361
Query: 400 IWNAIRFGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVIL 459
IWNAIRFGTVLENVVFDEH RE DYSD+SVTE +RAAYPIE IPNAKLPCVGPHP
Sbjct: 362 IWNAIRFGTVLENVVFDEHDREADYSDQSVTEESRAAYPIECIPNAKLPCVGPHPKTAYY 541
Query: 460 L 460
L
Sbjct: 542 L 544
>AW351069 homologue to GP|13785467|db phosphoenolpyruvate carboxykinase
{Flaveria trinervia}, partial (22%)
Length = 586
Score = 269 bits (687), Expect = 3e-72
Identities = 127/134 (94%), Positives = 127/134 (94%)
Frame = -2
Query: 360 DHNRYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFGTVLENVVFDEHT 419
DHNRYLIGDDEH W D GVSNIEGGCYAKCI SREKEPDIWNAIRFGTVLENVVFDEH
Sbjct: 585 DHNRYLIGDDEHXWGDNGVSNIEGGCYAKCIXXSREKEPDIWNAIRFGTVLENVVFDEHD 406
Query: 420 REVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVILLACDAFGVLPPVSKLNLAQT 479
REVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHP NVILLACDAFGVLPPVSKLNLAQT
Sbjct: 405 REVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPKNVILLACDAFGVLPPVSKLNLAQT 226
Query: 480 MYHFISGYTALVAG 493
MYHFISGYTALVAG
Sbjct: 225 MYHFISGYTALVAG 184
Score = 69.3 bits (168), Expect = 5e-12
Identities = 32/35 (91%), Positives = 34/35 (96%)
Frame = -2
Query: 617 LAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
+AGLFKNNFETFTNYKIGKG+NNLTEEILAAGP F
Sbjct: 192 VAGLFKNNFETFTNYKIGKGNNNLTEEILAAGPIF 88
>BU546503
Length = 598
Score = 183 bits (465), Expect(2) = 2e-67
Identities = 88/103 (85%), Positives = 94/103 (90%)
Frame = -2
Query: 503 ATFSACFGAAFIMLHPTKYAAMLAEKMEQHGATGWLVNTGWSGGSYGSGSRIKLQYTRKI 562
ATFSACFGAAFIMLHPTKYAAML EKM++HG WLVNTGWS GSYG+GSRIKLQYTRKI
Sbjct: 597 ATFSACFGAAFIMLHPTKYAAMLXEKMQKHGXXXWLVNTGWSAGSYGNGSRIKLQYTRKI 418
Query: 563 IDAIHSGSLLKAEFQKTPIFGLEIPTEVEGVPSEILDPVNTWS 605
IDAIHSGS L AE++KT IFGLEIPTEVEGVPSEILDP+NT S
Sbjct: 417 IDAIHSGSXLNAEYKKTEIFGLEIPTEVEGVPSEILDPMNTVS 289
Score = 91.7 bits (226), Expect(2) = 2e-67
Identities = 43/48 (89%), Positives = 46/48 (95%)
Frame = -1
Query: 604 WSDKDAYQETLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
WS+K AY+ETLLKLAGLFKNNFETFTNYKIGKG+NNLTEEILAAGP F
Sbjct: 205 WSNKKAYKETLLKLAGLFKNNFETFTNYKIGKGNNNLTEEILAAGPIF 62
>BQ785985
Length = 421
Score = 241 bits (614), Expect = 1e-63
Identities = 110/127 (86%), Positives = 122/127 (95%)
Frame = +1
Query: 215 LDKVFVNDQFLNWDPENRIKVRIVAARAYHSLFMHNMCIRPTPEELENFGTPDFTIYNAG 274
+ +V+VNDQFLNWDPE+RIKVRIV+ARAYHSLFMHNMCIRPTP+ELE FGTPDFTIYNAG
Sbjct: 40 IHQVYVNDQFLNWDPEHRIKVRIVSARAYHSLFMHNMCIRPTPKELEEFGTPDFTIYNAG 219
Query: 275 QFPCNRFTHYMTSSTSVDLNLARREMVILGTQYAGEMKKGLFGVMHYLMPKRKILSLHSG 334
FPCNR+THYMTSSTS+D+N+AR+EMVILGTQYAGEMKKGLFG+MHYLMPKR ILSLHSG
Sbjct: 220 LFPCNRYTHYMTSSTSIDINIARKEMVILGTQYAGEMKKGLFGLMHYLMPKRYILSLHSG 399
Query: 335 CNMGKDG 341
NMGKDG
Sbjct: 400 SNMGKDG 420
>TC204275 homologue to UP|PPCK_CUCSA (P42066) Phosphoenolpyruvate
carboxykinase [ATP] (PEP carboxykinase)
(Phosphoenolpyruvate carboxylase) (PEPCK) , partial
(30%)
Length = 611
Score = 154 bits (390), Expect = 9e-38
Identities = 71/81 (87%), Positives = 78/81 (95%)
Frame = +1
Query: 171 RDKRVVRDAVTEDDLWWGKGSPNIEMDEQTFLVNRERAVDYLNSLDKVFVNDQFLNWDPE 230
RDKRVV+D +T++DLWWGKGSPNIEMDE +F+VNRERAVDYLNSLDKVFVNDQFLNWD E
Sbjct: 7 RDKRVVKDDLTKNDLWWGKGSPNIEMDEHSFMVNRERAVDYLNSLDKVFVNDQFLNWDTE 186
Query: 231 NRIKVRIVAARAYHSLFMHNM 251
NRIKVRIV+ARAYHSLFMHNM
Sbjct: 187 NRIKVRIVSARAYHSLFMHNM 249
Score = 135 bits (341), Expect(2) = 3e-61
Identities = 64/68 (94%), Positives = 66/68 (96%)
Frame = +3
Query: 408 TVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAKLPCVGPHPTNVILLACDAFGV 467
+VLENVVFDEH REVDYSDKSVTENTRAAYPI+YIPNAKLPCVGPHP NVILLACDAFGV
Sbjct: 408 SVLENVVFDEHDREVDYSDKSVTENTRAAYPIKYIPNAKLPCVGPHPKNVILLACDAFGV 587
Query: 468 LPPVSKLN 475
LPPVSKLN
Sbjct: 588 LPPVSKLN 611
Score = 119 bits (297), Expect(2) = 3e-61
Identities = 63/105 (60%), Positives = 70/105 (66%)
Frame = +2
Query: 303 LGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHN 362
+GTQ E K GL+ +P+ L++H C + TGKTTLSTDHN
Sbjct: 173 IGTQRT-ESKSGLY------LPE---LTIHCSCT-------------TCTGKTTLSTDHN 283
Query: 363 RYLIGDDEHCWSDKGVSNIEGGCYAKCIDLSREKEPDIWNAIRFG 407
RYLIGDDEHCW D GVSNIEGGCYAKCIDLSREKEPDIWNAI G
Sbjct: 284 RYLIGDDEHCWGDNGVSNIEGGCYAKCIDLSREKEPDIWNAISVG 418
>CA851920 similar to GP|13785471|dbj phosphoenolpyruvate carboxykinase
{Flaveria pringlei}, partial (13%)
Length = 494
Score = 150 bits (380), Expect = 1e-36
Identities = 79/99 (79%), Positives = 86/99 (86%), Gaps = 1/99 (1%)
Frame = +1
Query: 552 SRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLEIPTEVEGVPSEILDPVNTWSDKDAYQ 611
SRIKL YTRKIIDAIHSGSLL E++KT IFGLEIPTEVEGVPSEIL+P NTWSDK AY+
Sbjct: 4 SRIKLSYTRKIIDAIHSGSLLDVEYKKTEIFGLEIPTEVEGVPSEILEPENTWSDKQAYK 183
Query: 612 ETLLKLAGLFKNNFETFTNYKIGKGDNN-LTEEILAAGP 649
ETLLKLAGLFKNNFETFT G+NN +T+EILAAGP
Sbjct: 184 ETLLKLAGLFKNNFETFT-----YGENNQVTKEILAAGP 285
>BI320935 similar to GP|13785471|dbj phosphoenolpyruvate carboxykinase
{Flaveria pringlei}, partial (13%)
Length = 303
Score = 147 bits (371), Expect = 1e-35
Identities = 74/92 (80%), Positives = 79/92 (85%)
Frame = +2
Query: 535 TGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLEIPTEVEGVP 594
+ WLVNTGWS GSYG GSRIKLQYTRKIIDAIHSGSLL AE++KT IFGLEIPTEVEGVP
Sbjct: 26 SSWLVNTGWSAGSYGYGSRIKLQYTRKIIDAIHSGSLLNAEYKKTEIFGLEIPTEVEGVP 205
Query: 595 SEILDPVNTWSDKDAYQETLLKLAGLFKNNFE 626
SEILDPVNT S++ LLKLAGL KNNFE
Sbjct: 206 SEILDPVNTCSNQHG*GGGLLKLAGLLKNNFE 301
>BE554863
Length = 374
Score = 145 bits (366), Expect = 6e-35
Identities = 69/77 (89%), Positives = 74/77 (95%)
Frame = +3
Query: 510 GAAFIMLHPTKYAAMLAEKMEQHGATGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSG 569
GAAFIMLHPTKYAAMLAEKM++HGATGWLVNTGWS GSYG+GSRIKLQYTRKIIDAIHSG
Sbjct: 144 GAAFIMLHPTKYAAMLAEKMQKHGATGWLVNTGWSAGSYGNGSRIKLQYTRKIIDAIHSG 323
Query: 570 SLLKAEFQKTPIFGLEI 586
SLL AE++KT IFGLEI
Sbjct: 324 SLLNAEYKKTEIFGLEI 374
>TC204272 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase ,
partial (16%)
Length = 513
Score = 143 bits (360), Expect = 3e-34
Identities = 75/104 (72%), Positives = 84/104 (80%), Gaps = 4/104 (3%)
Frame = +2
Query: 5 NGNGIATNGLPSIHTQK--NGICHDDSVPTVKANTIDELHSLQKKKSAPGTPISGTQTPF 62
NGNG A NGL I TQK +GICHDDS PTVKA TIDELHSLQKKKSAP TP++G Q PF
Sbjct: 194 NGNGSARNGLAKIQTQKKHDGICHDDSGPTVKAQTIDELHSLQKKKSAPTTPVTGFQAPF 373
Query: 63 T--SDPERQQQQLQSISASLASLTRETGPKVVKGDPAKKLENPK 104
++ ER +QQLQSISASLASLTRETGPK+VKGDPA+K + K
Sbjct: 374 ATLTEEERHKQQLQSISASLASLTRETGPKLVKGDPARKSDTQK 505
>BQ453291
Length = 414
Score = 80.9 bits (198), Expect(2) = 4e-33
Identities = 39/43 (90%), Positives = 41/43 (94%)
Frame = +3
Query: 609 AYQETLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
AY+ETLLKLAGLFKNNF TFTNYKIGKG+NNLTEEILAAGP F
Sbjct: 132 AYKETLLKLAGLFKNNFFTFTNYKIGKGNNNLTEEILAAGPIF 260
Score = 79.7 bits (195), Expect(2) = 4e-33
Identities = 37/43 (86%), Positives = 41/43 (95%)
Frame = +1
Query: 565 AIHSGSLLKAEFQKTPIFGLEIPTEVEGVPSEILDPVNTWSDK 607
AIHSGSLL AE++KT IFGLEIPTEVEGVPSEILDP+NTWS+K
Sbjct: 1 AIHSGSLLNAEYKKTEIFGLEIPTEVEGVPSEILDPMNTWSNK 129
>BU081591 homologue to SP|Q9SLZ0|PPCK_ Phosphoenolpyruvate carboxykinase
[ATP] (EC 4.1.1.49) (PEP carboxykinase), partial (10%)
Length = 426
Score = 132 bits (333), Expect = 4e-31
Identities = 72/122 (59%), Positives = 81/122 (66%), Gaps = 35/122 (28%)
Frame = +1
Query: 475 NLAQTMYHFISGYTALV----------------------------------AGTEDGIKE 500
NLAQTMYHFISGYTALV AGTE+GIKE
Sbjct: 1 NLAQTMYHFISGYTALVMFHINLASFVLTIF*SCFVSSVILTILYL**L*VAGTEEGIKE 180
Query: 501 PQATFSACFGAAFIMLHPTKYAAMLAEKMEQHGATGWLVNTGWSGGSYGSGS-RIKLQYT 559
PQATFSACFGAAFIMLHPTKYAAMLAEKM++HGATGWLVNTGWS G Y + S +I++++
Sbjct: 181 PQATFSACFGAAFIMLHPTKYAAMLAEKMQKHGATGWLVNTGWSAGRYINLSLKIEIEFL 360
Query: 560 RK 561
K
Sbjct: 361 SK 366
>TC204269 homologue to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase
, partial (11%)
Length = 461
Score = 87.0 bits (214), Expect = 2e-17
Identities = 44/55 (80%), Positives = 47/55 (85%), Gaps = 2/55 (3%)
Frame = +2
Query: 312 KKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDH--NRY 364
+KGLFGVMHYLMPKR+ILSLHSGCNMGKDGDVALFFGLSG +S DH NRY
Sbjct: 5 EKGLFGVMHYLMPKRQILSLHSGCNMGKDGDVALFFGLSG----IVSADHRLNRY 157
Score = 78.6 bits (192), Expect = 8e-15
Identities = 33/35 (94%), Positives = 33/35 (94%)
Frame = +3
Query: 351 GTGKTTLSTDHNRYLIGDDEHCWSDKGVSNIEGGC 385
GTGKTTLSTDHNRYLIGDDEHCW D GVSNIEGGC
Sbjct: 357 GTGKTTLSTDHNRYLIGDDEHCWGDNGVSNIEGGC 461
>BE658836 homologue to GP|13785467|dbj phosphoenolpyruvate carboxykinase
{Flaveria trinervia}, partial (13%)
Length = 481
Score = 77.4 bits (189), Expect = 2e-14
Identities = 37/39 (94%), Positives = 38/39 (96%)
Frame = -2
Query: 613 TLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
TLLKLAGLFKNNFETFTNYKIGKG+NNLTEEILAAGP F
Sbjct: 255 TLLKLAGLFKNNFETFTNYKIGKGNNNLTEEILAAGPIF 139
Score = 71.2 bits (173), Expect = 1e-12
Identities = 32/35 (91%), Positives = 34/35 (96%)
Frame = -2
Query: 217 KVFVNDQFLNWDPENRIKVRIVAARAYHSLFMHNM 251
+VFVNDQFLNWD ENRIKVRIV+ARAYHSLFMHNM
Sbjct: 417 QVFVNDQFLNWDTENRIKVRIVSARAYHSLFMHNM 313
Score = 48.1 bits (113), Expect = 1e-05
Identities = 33/75 (44%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Frame = -3
Query: 303 LGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHN 362
+GTQ E K GL+ +P+ L++H C + TGKTTLSTDHN
Sbjct: 389 IGTQRT-ESKSGLY------LPE---LTIHCSCT-------------TCTGKTTLSTDHN 279
Query: 363 RYLIGDDEH--CWSD 375
RYLIGDDEH W D
Sbjct: 278 RYLIGDDEHY*SWLD 234
>AW156734 homologue to PIR|T05900|T059 phosphoenolpyruvate carboxykinase
(ATP) (EC 4.1.1.49) - Arabidopsis thaliana, partial
(14%)
Length = 391
Score = 77.4 bits (189), Expect = 2e-14
Identities = 37/39 (94%), Positives = 38/39 (96%)
Frame = +2
Query: 613 TLLKLAGLFKNNFETFTNYKIGKGDNNLTEEILAAGPNF 651
TLLKLAGLFKNNFETFTNYKIGKG+NNLTEEILAAGP F
Sbjct: 218 TLLKLAGLFKNNFETFTNYKIGKGNNNLTEEILAAGPIF 334
Score = 71.2 bits (173), Expect = 1e-12
Identities = 32/35 (91%), Positives = 34/35 (96%)
Frame = +2
Query: 217 KVFVNDQFLNWDPENRIKVRIVAARAYHSLFMHNM 251
+VFVNDQFLNWD ENRIKVRIV+ARAYHSLFMHNM
Sbjct: 56 QVFVNDQFLNWDTENRIKVRIVSARAYHSLFMHNM 160
Score = 48.1 bits (113), Expect = 1e-05
Identities = 33/75 (44%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Frame = +3
Query: 303 LGTQYAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHN 362
+GTQ E K GL+ +P+ L++H C + TGKTTLSTDHN
Sbjct: 84 IGTQRT-ESKSGLY------LPE---LTIHCSCT-------------TCTGKTTLSTDHN 194
Query: 363 RYLIGDDEH--CWSD 375
RYLIGDDEH W D
Sbjct: 195 RYLIGDDEHY*SWLD 239
>AW156731 weakly similar to GP|13785467|dbj phosphoenolpyruvate carboxykinase
{Flaveria trinervia}, partial (8%)
Length = 184
Score = 72.8 bits (177), Expect = 5e-13
Identities = 37/60 (61%), Positives = 44/60 (72%)
Frame = +2
Query: 527 EKMEQHGATGWLVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSLLKAEFQKTPIFGLEI 586
+KM++ GAT W+ NTG S GSYG+G R+KL Y KIIDAIH G LL A + KT I GLEI
Sbjct: 2 KKMQKRGATVWIENTGRSVGSYGNGIRMKLLYIWKIIDAIHYGGLLTA*YNKTEIIGLEI 181
>AW279385
Length = 283
Score = 69.3 bits (168), Expect = 5e-12
Identities = 40/94 (42%), Positives = 52/94 (54%), Gaps = 1/94 (1%)
Frame = -1
Query: 479 TMYHFISGYTALVAGTEDGIKEPQATFSACFGAAF-IMLHPTKYAAMLAEKMEQHGATGW 537
+MYH I + VA T A SA F ++LH + +++ + ATGW
Sbjct: 283 SMYHVIYSISRYVAPTPRRH*HASAISSASFSRNHSVLLHRPNDSTNASQQSHKQVATGW 104
Query: 538 LVNTGWSGGSYGSGSRIKLQYTRKIIDAIHSGSL 571
LV TGWS S+G+G RI LQYT+ I D IHSGSL
Sbjct: 103 LVTTGWSATSHGNGIRINLQYTKLISDPIHSGSL 2
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,065,420
Number of Sequences: 63676
Number of extensions: 375663
Number of successful extensions: 2012
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1955
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2004
length of query: 651
length of database: 12,639,632
effective HSP length: 103
effective length of query: 548
effective length of database: 6,081,004
effective search space: 3332390192
effective search space used: 3332390192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0177.1


