

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
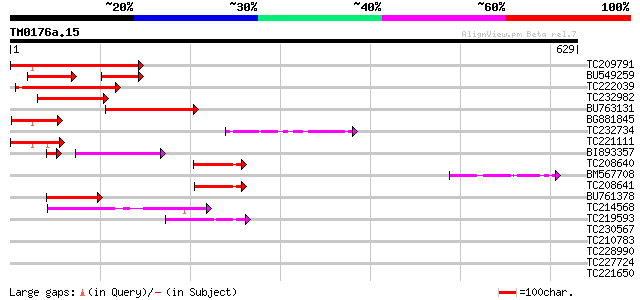
Query= TM0176a.15
(629 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209791 homologue to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, p... 237 1e-62
BU549259 87 5e-35
TC222039 118 7e-27
TC232982 99 6e-21
BU763131 80 3e-15
BG881845 similar to GP|9759604|dbj| contains similarity to cycli... 68 1e-11
TC232734 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 67 2e-11
TC221111 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 64 2e-10
BI893357 similar to GP|13924511|gb ania-6a type cyclin {Arabidop... 57 4e-09
TC208640 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 59 9e-09
BM567708 57 3e-08
TC208641 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 56 6e-08
BU761378 49 9e-06
TC214568 cyclin H [Glycine max] 46 6e-05
TC219593 similar to UP|Q9FJK6 (Q9FJK6) Cyclin C-like protein, pa... 45 1e-04
TC230567 similar to UP|Q9FGU0 (Q9FGU0) Arabidopsis thaliana geno... 38 0.012
TC210783 weakly similar to UP|O14760 (O14760) Small intestinal m... 37 0.020
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 34 0.22
TC227724 32 1.1
TC221650 similar to UP|Q6S4P4 (Q6S4P4) BZIP transcription factor... 31 1.5
>TC209791 homologue to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (20%)
Length = 1048
Score = 237 bits (605), Expect = 1e-62
Identities = 119/152 (78%), Positives = 130/152 (85%), Gaps = 4/152 (2%)
Frame = +3
Query: 1 MAGLLLGDTSHDGTHQSGSQGCS----QENGSRWYLSRKEIEENSPSKQDGVDLKKEAYL 56
MAGL+ G+ SH GT S S +E+ RWY+SRKEIEE+SPS++DG+DLKKE YL
Sbjct: 153 MAGLMPGELSHHGTSDGNSGKSSIEKQEESLGRWYMSRKEIEEHSPSRKDGIDLKKETYL 332
Query: 57 RKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEET 116
RKSYCT+LQDLGMRLKVPQVTIA+AIIFCHRFFLRQSHAKNDRR IAT CMFLAGKVEET
Sbjct: 333 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEET 512
Query: 117 PRPLKDVILVSYEIIHKKDPAAVQRIKQKEVY 148
PRPLKDVILVSYEIIHKKDPAA QRIKQK Y
Sbjct: 513 PRPLKDVILVSYEIIHKKDPAAAQRIKQKVFY 608
>BU549259
Length = 651
Score = 87.4 bits (215), Expect(2) = 5e-35
Identities = 43/47 (91%), Positives = 43/47 (91%)
Frame = -1
Query: 102 IATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVY 148
IAT CMFLAGKVEETPRPLKDVILVSYEIIHKKDPAA QRIKQK Y
Sbjct: 486 IATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAAQRIKQKVFY 346
Score = 79.3 bits (194), Expect(2) = 5e-35
Identities = 38/55 (69%), Positives = 44/55 (79%)
Frame = -2
Query: 20 QGCSQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVP 74
Q +E+ R Y SRKEIE SPS++DG+DLKKE LRKSYCT+LQDLGMRLKVP
Sbjct: 650 QDKQEESLGRXYXSRKEIEXXSPSRKDGIDLKKETXLRKSYCTFLQDLGMRLKVP 486
>TC222039
Length = 684
Score = 118 bits (296), Expect = 7e-27
Identities = 59/118 (50%), Positives = 84/118 (71%), Gaps = 1/118 (0%)
Frame = +2
Query: 7 GDTSHDGTHQSGSQGCSQ-ENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQ 65
GD S T S + CS E+ ++SR EI+ +SPS++DG+D+ E +LR SYC +LQ
Sbjct: 338 GDAS---TSASCKRDCSIFEDDKPVFMSRDEIDRHSPSRKDGIDVHHETHLRYSYCAFLQ 508
Query: 66 DLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDV 123
+LGMRL++PQ I +A++ CHRFF+RQSHA +DR +IAT+ +FL K EE PRPL ++
Sbjct: 509 NLGMRLELPQNIIGTAMVLCHRFFVRQSHACHDRFLIATAALFLTAKSEEAPRPLNNI 682
>TC232982
Length = 589
Score = 99.0 bits (245), Expect = 6e-21
Identities = 43/79 (54%), Positives = 64/79 (80%)
Frame = +3
Query: 31 YLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFL 90
++SR EI+ SPS++DG+D+ +E +LR SYC +LQ+LGM L +PQ TI +A++ CHRFF+
Sbjct: 351 FMSRDEIDRCSPSRKDGIDVLRERHLRYSYCAFLQNLGMWLNLPQTTIGTAMVLCHRFFV 530
Query: 91 RQSHAKNDRRIIATSCMFL 109
R+SHA +DR +IAT+ +FL
Sbjct: 531 RRSHACHDRFLIATAALFL 587
>BU763131
Length = 421
Score = 80.1 bits (196), Expect = 3e-15
Identities = 40/103 (38%), Positives = 64/103 (61%)
Frame = +1
Query: 107 MFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATL 166
+FL K E+ PR L +V+ S EI++K+D A + + +EQ +E +L E+++L TL
Sbjct: 1 LFLTAKSEKAPRHLNNVLRTSSEILYKQDFALLSYRFPVDWFEQYRERVLEAEQLILTTL 180
Query: 167 GFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSL 209
F+ +VQHPY PL + KL +S+ L +A N V++G+ T L
Sbjct: 181 NFELNVQHPYVPLTSVLNKLGLSKTVLVNLALNLVSEGIFTRL 309
>BG881845 similar to GP|9759604|dbj| contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(5%)
Length = 410
Score = 67.8 bits (164), Expect = 1e-11
Identities = 34/60 (56%), Positives = 42/60 (69%), Gaps = 4/60 (6%)
Frame = +3
Query: 3 GLLLGDTSHDGTHQSGSQGCSQ----ENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRK 58
GL+ G+ H GT S SQ E+ RWY+SRKEIEE+SPS++DG+DLKKE YLRK
Sbjct: 231 GLMPGELPHHGTPDGNSGKSSQDKQEESLGRWYMSRKEIEEHSPSRKDGIDLKKETYLRK 410
>TC232734 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (7%)
Length = 536
Score = 67.0 bits (162), Expect = 2e-11
Identities = 53/150 (35%), Positives = 80/150 (53%), Gaps = 3/150 (2%)
Frame = -1
Query: 240 QVFDVTLTAHQLEEVSNQMLELYEQNQIAPS--NDVEGTAGGGNRATPKASTSNDEAATT 297
Q FDVT QLEEVSNQMLELYEQN++ P+ ++VEG+A GG RA KA T+N++ A+
Sbjct: 536 QEFDVT--PRQLEEVSNQMLELYEQNRLPPAQGSEVEGSA-GGTRAASKAPTANEDQASK 366
Query: 298 NNNLYIRGPSPRLEASKLAISKNVVSSANHVGPVSNHGTNGSTEM-IHLVEGDAKGNQHP 356
I +P+ ++ + VV N +GS EM + + + +
Sbjct: 365 Q----ISSQAPQ----HSSVERTVVPQRG----TENQSNDGSAEMGSDITDHNLDIRESH 222
Query: 357 EQEPPPHKENLQEAQDTVRSRSGYSEEPEI 386
E H++N +E + RS+SG + +I
Sbjct: 221 NSEQLTHQDNKREVSN--RSKSGTERDQDI 138
>TC221111 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (3%)
Length = 557
Score = 64.3 bits (155), Expect = 2e-10
Identities = 37/66 (56%), Positives = 45/66 (68%), Gaps = 6/66 (9%)
Frame = +2
Query: 2 AGLLLGDTSHDGTHQSGSQGCS---QENGSRWYLSRKEIEEN---SPSKQDGVDLKKEAY 55
AGLL+GD S GT Q SQ S E+GSRWY SRK+IE +PS+QDG+DLKK+ +
Sbjct: 191 AGLLVGDVSCHGTCQGISQRYS*EKPEDGSRWYFSRKKIERKLTMTPSEQDGIDLKKDTH 370
Query: 56 LRKSYC 61
L KS C
Sbjct: 371 LCKS*C 388
>BI893357 similar to GP|13924511|gb ania-6a type cyclin {Arabidopsis
thaliana}, partial (33%)
Length = 421
Score = 57.0 bits (136), Expect(2) = 4e-09
Identities = 30/101 (29%), Positives = 58/101 (56%), Gaps = 1/101 (0%)
Frame = +2
Query: 74 PQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHK 133
PQ +A+ + HRF+ ++S A+ + + +A SC++LA K+EE PR + VI+V + + +
Sbjct: 119 PQAVMATGQVLFHRFYCKKSFARFNVKKVAASCVWLASKLEENPRKARQVIIVFHRMECR 298
Query: 134 KDPAAVQRIK-QKEVYEQQKELILLGERVVLATLGFDFHVQ 173
++ ++ + + Y K + ER +L +GF HV+
Sbjct: 299 REDFPMEHLDLYSKKYVDLKMELSRTERHILKEMGFICHVE 421
Score = 22.3 bits (46), Expect(2) = 4e-09
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = +3
Query: 41 SPSKQDGVDLKKEAYLR 57
SPS++DG+D E LR
Sbjct: 60 SPSRKDGIDEATETSLR 110
>TC208640 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (4%)
Length = 600
Score = 58.5 bits (140), Expect = 9e-09
Identities = 33/60 (55%), Positives = 42/60 (70%), Gaps = 2/60 (3%)
Frame = +3
Query: 205 LRTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLW--WQVFDVTLTAHQLEEVSNQMLELY 262
LR+SL LQFKP HIAAGA +LAAKFLN+ L WQ F T + L++VS Q++EL+
Sbjct: 156 LRSSLWLQFKPHHIAAGAAYLAAKFLNMDLAAYQNIWQEFQTTPSI--LQDVSQQLMELF 329
>BM567708
Length = 428
Score = 56.6 bits (135), Expect = 3e-08
Identities = 44/126 (34%), Positives = 69/126 (53%), Gaps = 3/126 (2%)
Frame = +2
Query: 489 RELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHL-VKPSHETD 547
RELEDGIELA + EKNK++ +++ SKP + G++ + + + HL +K + D
Sbjct: 8 RELEDGIELAVEDEKNKRERRQNWSKPDGEN-----HHGENHEETRDGRHLSMKGQFQKD 172
Query: 548 LSA--VEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGKKRHNYSFGSTHHNRIDYIE 605
+ EEGE+ +D D +N KR+ S P + E KKR S++HN D E
Sbjct: 173 MDEDNAEEGEM--ID--DASSSLNNRKRRMGSPPGRQPEMKKR----LDSSYHN--DLAE 322
Query: 606 DQNNVN 611
+N++N
Sbjct: 323 *RNSLN 340
>TC208641 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (4%)
Length = 513
Score = 55.8 bits (133), Expect = 6e-08
Identities = 32/59 (54%), Positives = 40/59 (67%), Gaps = 2/59 (3%)
Frame = +2
Query: 206 RTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLW--WQVFDVTLTAHQLEEVSNQMLELY 262
R+SL LQFKP HIAAGA +LAAKFLN+ L WQ F T + L+ VS Q++EL+
Sbjct: 8 RSSLWLQFKPHHIAAGAAYLAAKFLNMDLAAYQNIWQEFQTTPSI--LQAVSQQLMELF 178
>BU761378
Length = 434
Score = 48.5 bits (114), Expect = 9e-06
Identities = 22/63 (34%), Positives = 40/63 (62%)
Frame = +1
Query: 41 SPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRR 100
SPS++DG+D E LR C +Q+ G+ L++PQ +A+ + HRF+ ++S A+ + +
Sbjct: 241 SPSRKDGIDEATETSLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFYCKKSFARFNVK 420
Query: 101 IIA 103
+A
Sbjct: 421 KVA 429
>TC214568 cyclin H [Glycine max]
Length = 1214
Score = 45.8 bits (107), Expect = 6e-05
Identities = 46/191 (24%), Positives = 82/191 (42%), Gaps = 10/191 (5%)
Frame = +1
Query: 43 SKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRII 102
S+ + +++E ++ Y LQ++ + P A+A+I+ RF+L+ S ++ + I
Sbjct: 679 SRTKPLTIEEEQCIKVFYENKLQEVCNNFRFPHKIQATALIYFKRFYLQWSVMEHQPKHI 858
Query: 103 ATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVV 162
+C++ A K+EE VS E + K Q ++IL E +V
Sbjct: 859 MLTCIYAACKIEENH--------VSAEELGKG-------------ISQDHQMILNNEMIV 975
Query: 163 LATLGFDFHVQHPYKPLVEAIKKLKVSQNA----------LAQVAWNFVNDGLRTSLCLQ 212
+L FD V PY+ + I ++ NA L + A V+ + T L
Sbjct: 976 YQSLEFDLIVYAPYRSVEGFINDMEEFFNAGDNQLEMLKTLQETARFEVDKMMLTDAPLL 1155
Query: 213 FKPQHIAAGAI 223
F P +A A+
Sbjct: 1156FPPXQLALAAL 1188
>TC219593 similar to UP|Q9FJK6 (Q9FJK6) Cyclin C-like protein, partial (45%)
Length = 783
Score = 45.1 bits (105), Expect = 1e-04
Identities = 25/94 (26%), Positives = 47/94 (49%)
Frame = +1
Query: 174 HPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVK 233
HPY+ L ++ ++ + Q+ W VND + L L P IA I++A+ L K
Sbjct: 31 HPYRSLSPLLQDAGLNDLNMTQLTWGLVNDTYKMDLILVHPPHLIALACIYIAS-VLREK 207
Query: 234 LEKLWWQVFDVTLTAHQLEEVSNQMLELYEQNQI 267
W++ V + ++ +S ++L+ YE N++
Sbjct: 208 DTTAWFEELRVDMNV--VKNISMEILDFYESNRM 303
>TC230567 similar to UP|Q9FGU0 (Q9FGU0) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K21L19, partial (9%)
Length = 1418
Score = 38.1 bits (87), Expect = 0.012
Identities = 50/218 (22%), Positives = 89/218 (39%), Gaps = 5/218 (2%)
Frame = +3
Query: 311 EASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEA 370
+ + L +SK S N GP + G GS GD + +H E H+ + ++
Sbjct: 21 DLNSLGLSKR--SQENQSGPTNEKGLKGS--------GDEERAEH---EISGHRLSRKQR 161
Query: 371 QDTVRSRSGYSEEPEINIGRSNMK-----EDVELNDKHSSKNLNHRDGTFAPPSLEAIKK 425
+D S E+ + GRS ++ ++ + N SS +L +D
Sbjct: 162 EDM----SSDDEQQDSRRGRSKLERWTSHKERDFNVNKSSSSLKFKD------------- 290
Query: 426 IDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLI 485
ID+D A+ E K A K + + L R+ D A ++++ D +
Sbjct: 291 IDKDNNDASSEAGKPAY-EPAKTVDADNQHILSVEARDSADMENRDADTKESGDRHLDTV 467
Query: 486 ERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFEN 523
ER+++ + +L SEK +K S+P+ + EN
Sbjct: 468 ERLKKRSERFKLPMPSEKEALVIKKLESEPLPSAKSEN 581
>TC210783 weakly similar to UP|O14760 (O14760) Small intestinal mucin MUC3
(Fragment), partial (5%)
Length = 735
Score = 37.4 bits (85), Expect = 0.020
Identities = 37/131 (28%), Positives = 64/131 (48%), Gaps = 7/131 (5%)
Frame = +3
Query: 424 KKIDRDKVKAALEKR---KKAVGNITKKTELMDDEDLFERVRE--LEDGIELAAQSEKNK 478
K ++ + +K + K KKAVG +K E + E +E EDG++ A K
Sbjct: 321 KVLENNGIKKNIVKEIEDKKAVG--VEKVEEDKKNGVVEDAKEDKKEDGVKEA------K 476
Query: 479 QDDEDLIERVRE--LEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEH 536
+D ++++E V+E EDG+E A E K+DG + + + + + +++ K D +E
Sbjct: 477 EDKKEVVEEVKEDNKEDGVEEA--KENKKEDGVEEVKEVKEVDGVKEIKEDKEVDGVKE- 647
Query: 537 LHLVKPSHETD 547
VK E D
Sbjct: 648 ---VKEDKEVD 671
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 33.9 bits (76), Expect = 0.22
Identities = 26/79 (32%), Positives = 37/79 (45%)
Frame = +1
Query: 293 EAATTNNNLYIRGPSPRLEASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKG 352
E T + I P+ +SK AI+++ +SS+N +G T S E K
Sbjct: 2935 ERPTMREVVQILTELPKPPSSKHAITESSLSSSNSLGSP----TTASKE--------PKD 3078
Query: 353 NQHPEQEPPPHKENLQEAQ 371
NQHP Q PPP ++ AQ
Sbjct: 3079 NQHPPQSPPPDLLSI*SAQ 3135
>TC227724
Length = 799
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/71 (29%), Positives = 33/71 (45%), Gaps = 12/71 (16%)
Frame = +2
Query: 439 KKAVGNITKKTELMDDEDLFERVR------------ELEDGIELAAQSEKNKQDDEDLIE 486
+ V IT+ EL+D+ + E + ELE +E+ AQ E+ D + IE
Sbjct: 266 RSIVDEITENIELIDEHCVAETAKANSDREVESLNKELESNLEILAQKEQEVTDIKTRIE 445
Query: 487 RVRELEDGIEL 497
+RE +EL
Sbjct: 446 AIRERLSELEL 478
>TC221650 similar to UP|Q6S4P4 (Q6S4P4) BZIP transcription factor RF2b,
partial (44%)
Length = 697
Score = 31.2 bits (69), Expect = 1.5
Identities = 28/125 (22%), Positives = 54/125 (42%), Gaps = 1/125 (0%)
Frame = +2
Query: 417 PPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELE-DGIELAAQSE 475
P L + ID + K L R+ A + +K + ++L +V+ L+ + L+AQ
Sbjct: 254 PDKLAELWTIDPKRAKRILANRQSAARSKERKARYI--QELERKVQTLQTEATTLSAQLT 427
Query: 476 KNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEE 535
++D L EL+ ++ Q + + ++L K V+R + H D
Sbjct: 428 LYQRDTTGLSTENTELKLRLQAMEQQAQLRDALNEALKKEVERLKVATGEMMSHTDSFNL 607
Query: 536 HLHLV 540
+HL+
Sbjct: 608 GMHLM 622
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,891,763
Number of Sequences: 63676
Number of extensions: 321600
Number of successful extensions: 1580
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1575
length of query: 629
length of database: 12,639,632
effective HSP length: 103
effective length of query: 526
effective length of database: 6,081,004
effective search space: 3198608104
effective search space used: 3198608104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0176a.15


