

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.14
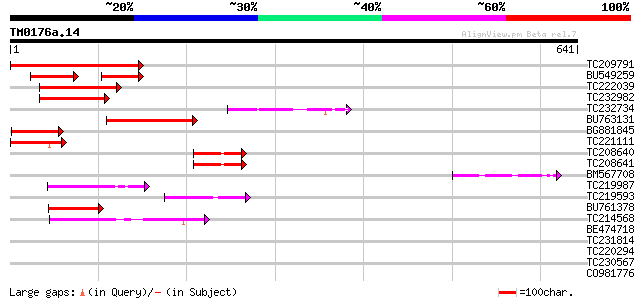
(641 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209791 homologue to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, p... 246 2e-65
BU549259 89 4e-36
TC222039 118 7e-27
TC232982 100 2e-21
TC232734 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 82 6e-16
BU763131 80 2e-15
BG881845 similar to GP|9759604|dbj| contains similarity to cycli... 76 5e-14
TC221111 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 73 4e-13
TC208640 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 69 8e-12
TC208641 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, par... 66 5e-11
BM567708 59 5e-09
TC219987 similar to UP|CG1C_ORYSA (P93411) G1/S-specific cyclin ... 56 6e-08
TC219593 similar to UP|Q9FJK6 (Q9FJK6) Cyclin C-like protein, pa... 53 4e-07
BU761378 50 2e-06
TC214568 cyclin H [Glycine max] 49 9e-06
BE474718 similar to GP|10177353|dbj cyclin C-like protein {Arabi... 39 0.005
TC231814 similar to UP|O02445 (O02445) LOX6 (Fragment), partial ... 36 0.046
TC220294 33 0.39
TC230567 similar to UP|Q9FGU0 (Q9FGU0) Arabidopsis thaliana geno... 33 0.39
CO981776 30 4.3
>TC209791 homologue to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (20%)
Length = 1048
Score = 246 bits (629), Expect = 2e-65
Identities = 123/152 (80%), Positives = 132/152 (85%), Gaps = 1/152 (0%)
Frame = +3
Query: 1 MAGLLLGDISHHGSYQSGSQGGSQDNQEYG-SRWYFSRKEIEENSPSKQDGVDLKKEAYL 59
MAGL+ G++SHHG+ S S + QE RWY SRKEIEE+SPS++DG+DLKKE YL
Sbjct: 153 MAGLMPGELSHHGTSDGNSGKSSIEKQEESLGRWYMSRKEIEEHSPSRKDGIDLKKETYL 332
Query: 60 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEET 119
RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRR IATVCMFLAGKVEET
Sbjct: 333 RKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRTIATVCMFLAGKVEET 512
Query: 120 PRPLKDVILVSYEIIHKKDPAAVQRIKQKDVY 151
PRPLKDVILVSYEIIHKKDPAA QRIKQK Y
Sbjct: 513 PRPLKDVILVSYEIIHKKDPAAAQRIKQKVFY 608
>BU549259
Length = 651
Score = 89.4 bits (220), Expect(2) = 4e-36
Identities = 44/47 (93%), Positives = 44/47 (93%)
Frame = -1
Query: 105 IATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVY 151
IATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAA QRIKQK Y
Sbjct: 486 IATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAAQRIKQKVFY 346
Score = 81.3 bits (199), Expect(2) = 4e-36
Identities = 41/55 (74%), Positives = 44/55 (79%), Gaps = 1/55 (1%)
Frame = -2
Query: 24 QDNQEYG-SRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVP 77
QD QE R Y SRKEIE SPS++DG+DLKKE LRKSYCTFLQDLGMRLKVP
Sbjct: 650 QDKQEESLGRXYXSRKEIEXXSPSRKDGIDLKKETXLRKSYCTFLQDLGMRLKVP 486
>TC222039
Length = 684
Score = 118 bits (296), Expect = 7e-27
Identities = 53/93 (56%), Positives = 72/93 (76%)
Frame = +2
Query: 34 YFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFL 93
+ SR EI+ +SPS++DG+D+ E +LR SYC FLQ+LGMRL++PQ I TA++ CHRFF+
Sbjct: 404 FMSRDEIDRHSPSRKDGIDVHHETHLRYSYCAFLQNLGMRLELPQNIIGTAMVLCHRFFV 583
Query: 94 RQSHAKNDRRIIATVCMFLAGKVEETPRPLKDV 126
RQSHA +DR +IAT +FL K EE PRPL ++
Sbjct: 584 RQSHACHDRFLIATAALFLTAKSEEAPRPLNNI 682
>TC232982
Length = 589
Score = 100 bits (249), Expect = 2e-21
Identities = 45/79 (56%), Positives = 62/79 (77%)
Frame = +3
Query: 34 YFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFL 93
+ SR EI+ SPS++DG+D+ +E +LR SYC FLQ+LGM L +PQ TI TA++ CHRFF+
Sbjct: 351 FMSRDEIDRCSPSRKDGIDVLRERHLRYSYCAFLQNLGMWLNLPQTTIGTAMVLCHRFFV 530
Query: 94 RQSHAKNDRRIIATVCMFL 112
R+SHA +DR +IAT +FL
Sbjct: 531 RRSHACHDRFLIATAALFL 587
>TC232734 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (7%)
Length = 536
Score = 82.4 bits (202), Expect = 6e-16
Identities = 59/145 (40%), Positives = 80/145 (54%), Gaps = 5/145 (3%)
Frame = -1
Query: 247 QEFDVTPRQLEEVSNQMLELYEQNRM--AQSNDVEGTTGGGNRATPKAPTSNDEAASTNR 304
QEFDVTPRQLEEVSNQMLELYEQNR+ AQ ++VEG + GG RA KAPT+N++ AS
Sbjct: 536 QEFDVTPRQLEEVSNQMLELYEQNRLPPAQGSEVEG-SAGGTRAASKAPTANEDQASKQI 360
Query: 305 NLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVED---DVKGNL 361
+ SS T P R N GS E+ + D D++ +
Sbjct: 359 SSQAPQHSSVERTVVPQ--------------RGTENQSNDGSAEMGSDITDHNLDIRESH 222
Query: 362 HPKQEPSAYEENMQEAQDMVRSRSG 386
+ +Q +++N +E + RS+SG
Sbjct: 221 NSEQ--LTHQDNKREVSN--RSKSG 159
Score = 29.6 bits (65), Expect = 4.3
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 4/73 (5%)
Frame = -1
Query: 383 SRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGR----SNIKEEDVEL 438
S++ A +Q KQ S + + +V R G E +SN G S+I + ++++
Sbjct: 404 SKAPTANEDQASKQISSQAPQHSSVERTVVPQR---GTENQSNDGSAEMGSDITDHNLDI 234
Query: 439 NDKHSSKNLNHRD 451
+ H+S+ L H+D
Sbjct: 233 RESHNSEQLTHQD 195
>BU763131
Length = 421
Score = 80.5 bits (197), Expect = 2e-15
Identities = 40/103 (38%), Positives = 63/103 (60%)
Frame = +1
Query: 110 MFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATL 169
+FL K E+ PR L +V+ S EI++K+D A + D +EQ +E +L E+++L TL
Sbjct: 1 LFLTAKSEKAPRHLNNVLRTSSEILYKQDFALLSYRFPVDWFEQYRERVLEAEQLILTTL 180
Query: 170 GFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSL 212
F+LNV HPY PL + K +++ L +A N V++G+ T L
Sbjct: 181 NFELNVQHPYVPLTSVLNKLGLSKTVLVNLALNLVSEGIFTRL 309
>BG881845 similar to GP|9759604|dbj| contains similarity to
cyclin~gene_id:K18C1.7 {Arabidopsis thaliana}, partial
(5%)
Length = 410
Score = 75.9 bits (185), Expect = 5e-14
Identities = 36/60 (60%), Positives = 44/60 (73%), Gaps = 1/60 (1%)
Frame = +3
Query: 3 GLLLGDISHHGSYQSGSQGGSQDNQEYG-SRWYFSRKEIEENSPSKQDGVDLKKEAYLRK 61
GL+ G++ HHG+ S SQD QE RWY SRKEIEE+SPS++DG+DLKKE YLRK
Sbjct: 231 GLMPGELPHHGTPDGNSGKSSQDKQEESLGRWYMSRKEIEEHSPSRKDGIDLKKETYLRK 410
>TC221111 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (3%)
Length = 557
Score = 72.8 bits (177), Expect = 4e-13
Identities = 38/66 (57%), Positives = 48/66 (72%), Gaps = 3/66 (4%)
Frame = +2
Query: 2 AGLLLGDISHHGSYQSGSQGGSQDNQEYGSRWYFSRKEIEEN---SPSKQDGVDLKKEAY 58
AGLL+GD+S HG+ Q SQ S + E GSRWYFSRK+IE +PS+QDG+DLKK+ +
Sbjct: 191 AGLLVGDVSCHGTCQGISQRYS*EKPEDGSRWYFSRKKIERKLTMTPSEQDGIDLKKDTH 370
Query: 59 LRKSYC 64
L KS C
Sbjct: 371 LCKS*C 388
>TC208640 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (4%)
Length = 600
Score = 68.6 bits (166), Expect = 8e-12
Identities = 35/60 (58%), Positives = 45/60 (74%)
Frame = +3
Query: 208 LRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELY 267
LR+SL LQFKPHHIAAGA +LAAKFL + L + + WQEF TP L++VS Q++EL+
Sbjct: 156 LRSSLWLQFKPHHIAAGAAYLAAKFLNMDLAA--YQNIWQEFQTTPSILQDVSQQLMELF 329
>TC208641 similar to UP|Q9FKE6 (Q9FKE6) Similarity to cyclin, partial (4%)
Length = 513
Score = 65.9 bits (159), Expect = 5e-11
Identities = 34/59 (57%), Positives = 43/59 (72%)
Frame = +2
Query: 209 RTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELY 267
R+SL LQFKPHHIAAGA +LAAKFL + L + + WQEF TP L+ VS Q++EL+
Sbjct: 8 RSSLWLQFKPHHIAAGAAYLAAKFLNMDLAA--YQNIWQEFQTTPSILQAVSQQLMELF 178
>BM567708
Length = 428
Score = 59.3 bits (142), Expect = 5e-09
Identities = 46/127 (36%), Positives = 71/127 (55%), Gaps = 3/127 (2%)
Frame = +2
Query: 501 RELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHL-VKPSHEAD 559
RELEDGIELA + EKNK++ R++ SK + HG++ + + + HL +K + D
Sbjct: 8 RELEDGIELAVEDEKNKRERRQNWSKPDGEN-----HHGENHEETRDGRHLSMKGQFQKD 172
Query: 560 LSA--VEEGEVSALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGSGSTHHNRIDYIE 617
+ EEGE+ +D D +N+KR++ S P E KKR + S++HN D E
Sbjct: 173 MDEDNAEEGEM--ID--DASSSLNNRKRRMGSPPGRQPEMKKRLD----SSYHN--DLAE 322
Query: 618 DRNKVNR 624
RN +N+
Sbjct: 323 *RNSLNK 343
>TC219987 similar to UP|CG1C_ORYSA (P93411) G1/S-specific cyclin C-type,
partial (53%)
Length = 728
Score = 55.8 bits (133), Expect = 6e-08
Identities = 33/116 (28%), Positives = 63/116 (53%)
Frame = +1
Query: 43 NSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDR 102
N+ K+ G+ L+ ++ ++ L ++KV Q +ATA+ + R + R+S A+ D
Sbjct: 382 NTLDKEKGITLEDFKLIKMHMANYILKLAQQVKVRQRVVATAVTYMRRVYTRKSMAEYDP 561
Query: 103 RIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELI 158
R++A C++LA K EE+ ++ +LV Y I K R + KD+ E + +++
Sbjct: 562 RLVAPTCLYLASKAEES--TVQARLLVFY--IKKLYTDDKYRYEIKDILEMEMKIL 717
>TC219593 similar to UP|Q9FJK6 (Q9FJK6) Cyclin C-like protein, partial (45%)
Length = 783
Score = 53.1 bits (126), Expect = 4e-07
Identities = 28/97 (28%), Positives = 47/97 (47%)
Frame = +1
Query: 176 HHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKV 235
+HPY+ L ++ + + Q+ W VND + L L PH IA I++A+
Sbjct: 28 YHPYRSLSPLLQDAGLNDLNMTQLTWGLVNDTYKMDLILVHPPHLIALACIYIAS----- 192
Query: 236 KLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNRM 272
L W++E V ++ +S ++L+ YE NRM
Sbjct: 193 VLREKDTTAWFEELRVDMNVVKNISMEILDFYESNRM 303
>BU761378
Length = 434
Score = 50.4 bits (119), Expect = 2e-06
Identities = 23/63 (36%), Positives = 40/63 (62%)
Frame = +1
Query: 44 SPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRR 103
SPS++DG+D E LR C +Q+ G+ L++PQ +AT + HRF+ ++S A+ + +
Sbjct: 241 SPSRKDGIDEATETSLRIYGCDLIQESGILLRLPQAVMATGQVLFHRFYCKKSFARFNVK 420
Query: 104 IIA 106
+A
Sbjct: 421 KVA 429
>TC214568 cyclin H [Glycine max]
Length = 1214
Score = 48.5 bits (114), Expect = 9e-06
Identities = 48/191 (25%), Positives = 82/191 (42%), Gaps = 10/191 (5%)
Frame = +1
Query: 46 SKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRII 105
S+ + +++E ++ Y LQ++ + P ATA+I+ RF+L+ S ++ + I
Sbjct: 679 SRTKPLTIEEEQCIKVFYENKLQEVCNNFRFPHKIQATALIYFKRFYLQWSVMEHQPKHI 858
Query: 106 ATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVV 165
C++ A K+EE VS E + K Q ++IL E +V
Sbjct: 859 MLTCIYAACKIEENH--------VSAEELGKG-------------ISQDHQMILNNEMIV 975
Query: 166 LATLGFDLNVHHPYKPLVEAIKKFKVAQNA----------LAQVAWNFVNDGLRTSLCLQ 215
+L FDL V+ PY+ + I + NA L + A V+ + T L
Sbjct: 976 YQSLEFDLIVYAPYRSVEGFINDMEEFFNAGDNQLEMLKTLQETARFEVDKMMLTDAPLL 1155
Query: 216 FKPHHIAAGAI 226
F P +A A+
Sbjct: 1156FPPXQLALAAL 1188
>BE474718 similar to GP|10177353|dbj cyclin C-like protein {Arabidopsis
thaliana}, partial (28%)
Length = 420
Score = 39.3 bits (90), Expect = 0.005
Identities = 22/69 (31%), Positives = 34/69 (48%)
Frame = +3
Query: 204 VNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQM 263
VND + L L PH IA I++A+ L W++E V ++ +S ++
Sbjct: 207 VNDTYKMDLILVHPPHLIALACIYIASV-----LREKDTTAWFEELRVDMNVVKNISMEI 371
Query: 264 LELYEQNRM 272
L+ YE NRM
Sbjct: 372 LDFYESNRM 398
>TC231814 similar to UP|O02445 (O02445) LOX6 (Fragment), partial (7%)
Length = 620
Score = 36.2 bits (82), Expect = 0.046
Identities = 27/101 (26%), Positives = 50/101 (48%), Gaps = 7/101 (6%)
Frame = +2
Query: 519 DGRKSLSKSLDRSAFEN---MQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDID 575
+ R+ + S + F+N + H Q+ + L P+ E ++ E+G + +D+ +
Sbjct: 29 EARRGIEASEEEWHFKNGIGLTHKGAQEKPQIKFDLESPAEE-HIAESEDGGLEVIDETE 205
Query: 576 ----LGPKSSNQKRKVESSPDNFVEGKKRHNYGSGSTHHNR 612
LGP S N+++KVE +P G + SGS+H N+
Sbjct: 206 IELTLGPSSYNRRKKVE-TPLTSDSGHSLSSSSSGSSHVNK 325
>TC220294
Length = 1014
Score = 33.1 bits (74), Expect = 0.39
Identities = 31/165 (18%), Positives = 72/165 (42%), Gaps = 9/165 (5%)
Frame = +1
Query: 365 QEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQES 424
+E + +EE ++ +G + + +PS EE + + + G K ++
Sbjct: 37 EESTYFEEQPSGINSVLEDTNGSTETQEIGYDQPSFTEEIFEYLTNAQNNGEGDEKHKDD 216
Query: 425 NVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNI 484
+ N K D E D + ++ + N +G + ++ +R+ ++
Sbjct: 217 DAASCNTKARDEETIDNTQCQKMS-ETSKIEETNEDGYSSSLENNDESNKGERQIELVDV 393
Query: 485 SKKTELV-DDEDLIERVRELEDGIE-----LAAQSE---KNKQDG 520
SK++ + +D+DL+E+ + G++ ++A+ E KN +DG
Sbjct: 394 SKESNIASEDQDLLEKDQGKAIGLQQSTSCISAEEESTSKNWKDG 528
>TC230567 similar to UP|Q9FGU0 (Q9FGU0) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K21L19, partial (9%)
Length = 1418
Score = 33.1 bits (74), Expect = 0.39
Identities = 41/161 (25%), Positives = 67/161 (41%), Gaps = 17/161 (10%)
Frame = +3
Query: 392 QHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNI------KEEDVELNDKHSS- 444
+ + E S + ++ +DM S ++Q+S GRS + KE D +N SS
Sbjct: 111 ERAEHEISGHRLSRKQREDM----SSDDEQQDSRRGRSKLERWTSHKERDFNVNKSSSSL 278
Query: 445 ------KNLNHRDATFASPNLEGIKKIDRDKVKA-ALEKRKKAT-GNISKKTELVDDE-- 494
K+ N + P E K +D D ++E R A N T+ D
Sbjct: 279 KFKDIDKDNNDASSEAGKPAYEPAKTVDADNQHILSVEARDSADMENRDADTKESGDRHL 458
Query: 495 DLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFEN 535
D +ER+++ + +L SEK +K S+ L + EN
Sbjct: 459 DTVERLKKRSERFKLPMPSEKEALVIKKLESEPLPSAKSEN 581
>CO981776
Length = 714
Score = 29.6 bits (65), Expect = 4.3
Identities = 31/145 (21%), Positives = 61/145 (41%), Gaps = 9/145 (6%)
Frame = -1
Query: 465 IDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSL 524
I D K K+ + G ++T+ V E++ G+ + ++ KN+ + R+
Sbjct: 711 ISEDSDKXXKVKQLQLRGRTRRETKX-------RSVGEIQSGLRRSKRATKNRINYRQCE 553
Query: 525 SKSLDRSAFENMQHGKHQDHSE--EHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSN 582
+ + ++ + DHS+ E+ + S ++D S EE E+ D + N
Sbjct: 552 ASESETEFIKSEKSNSSADHSDPNENGEYMMESEDSDDSDNEEQEMKVDDPVTYPAVEEN 373
Query: 583 QK----RKVESSPDNFVE---GKKR 600
++ K+ S VE GK+R
Sbjct: 372 EQNQPPEKLSSPGQEEVESSTGKRR 298
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,438,141
Number of Sequences: 63676
Number of extensions: 302755
Number of successful extensions: 1419
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1380
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1408
length of query: 641
length of database: 12,639,632
effective HSP length: 103
effective length of query: 538
effective length of database: 6,081,004
effective search space: 3271580152
effective search space used: 3271580152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0176a.14


