

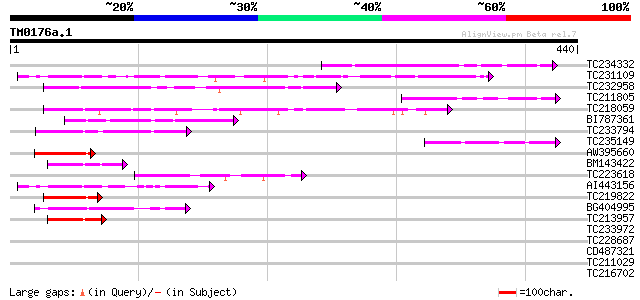
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.1
(440 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC234332 111 7e-25
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 87 1e-17
TC232958 79 4e-15
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 64 1e-10
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 63 2e-10
BI787361 59 4e-09
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 55 6e-08
TC235149 54 1e-07
AW395660 50 3e-06
BM143422 49 3e-06
TC223618 48 1e-05
AI443156 48 1e-05
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 48 1e-05
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 47 2e-05
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 44 1e-04
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 41 0.001
TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%) 41 0.001
CD487321 40 0.002
TC211029 similar to UP|Q96VI4 (Q96VI4) Protease 1, partial (3%) 40 0.002
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 38 0.010
>TC234332
Length = 644
Score = 111 bits (277), Expect = 7e-25
Identities = 68/183 (37%), Positives = 95/183 (51%)
Frame = +2
Query: 243 DGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVV 302
DG + GT+NW+ + S+ Y T+T++ L I S DL+ E+Y L P GL EVP
Sbjct: 17 DGASVRGTVNWLALPNSSSDYQWE-TVTIDDLVIFSYDLKNESYRYLLMPDGLLEVPHSP 193
Query: 303 PSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQ 362
P + VL CLC SH G HF W MK+FG KSWT+ L + Y L +H F H
Sbjct: 194 PELVVLKGCLCLSHRHGGNHFGFWLMKEFGVEKSWTRFLNISYDQLH---IHGGFLDHPV 364
Query: 363 LFPLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESL 422
+ LC+ + D ++L + ILY+KR NT E + ++ F S+ Y +S
Sbjct: 365 I--LCMSEDDGVVLLENGGH---GKFILYNKRDNTIECYGELDKGRF--QFLSYDYAQSF 523
Query: 423 VSP 425
V P
Sbjct: 524 VMP 532
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 87.4 bits (215), Expect = 1e-17
Identities = 102/382 (26%), Positives = 170/382 (43%), Gaps = 13/382 (3%)
Frame = +3
Query: 7 NTVTVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIK 66
N +TV+ M NLP +++ E+LS LPVKS+ RL+ CK W S II + F+
Sbjct: 63 NRITVVMMA-----NLPV-----EVVTEILSRLPVKSVIRLRSTCKWWRS-IIDSRHFVL 209
Query: 67 LHLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGD 126
HL++S L+L+ HR +HL L E + L+H + +
Sbjct: 210 FHLNKS----HSSLILR-------HR----SHLYSLDLKSPEQNPVELSHP---LMCYSN 335
Query: 127 RYHVVGCCNGLICLHGYSVDSIFQQGWLSFWN---------PATRLMSEKLGCFCVKTKN 177
V+G NGL+C+ + D ++ WN PA R + F +
Sbjct: 336 SIKVLGSSNGLLCISNVADD-------IALWNPFLRKHRILPADRFHRPQSSLFAARV-- 488
Query: 178 FDVKLSFGYDNSTDTYKVV----FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLP 233
FG+ + ++ YK++ F +++K S V ++TL + W+ N P +P
Sbjct: 489 ----YGFGHHSPSNDYKLLSITYFVDLQKRTFDSQVQLYTLKS----DSWK---NLPSMP 635
Query: 234 FSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPK 293
++ GV +SG+++W+ +R K P+ E I+S DL ET+ + P
Sbjct: 636 YAL-CCARTMGVFVSGSLHWLVTR-KLQPH--------EPDLIVSFDLTRETFHEVPLPV 785
Query: 294 GLDEVPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPL 353
++ V +L CLC + GT F +W M+ +G SW +L + + E +
Sbjct: 786 TVN--GDFDMQVALLGGCLCVV-EHRGTGFDVWVMRVYGSRNSWEKLFTLLENNDHHEMM 956
Query: 354 HSMFYCHYQLFPLCLHDGDTLI 375
S + + PL L DGD ++
Sbjct: 957 GSGKLKYVR--PLAL-DGDRVL 1013
>TC232958
Length = 758
Score = 79.0 bits (193), Expect = 4e-15
Identities = 74/236 (31%), Positives = 101/236 (42%), Gaps = 5/236 (2%)
Frame = +1
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
L DLI E+L LPVKSL R K VCKSW +ISD F K H A L +
Sbjct: 61 LPQDLITEILLRLPVKSLVRFKSVCKSW-LFLISDPRFAKSHFDLFAALADRILFIASSA 237
Query: 87 SHHDHREFCVT-HLSGSSLSL-VENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYS 144
+F + H +S+++ V+ P A P + ++G C G I LH S
Sbjct: 238 PELRSIDFNASLHDDSASVAVTVDLP----APKPYFHF-----VEIIGSCRGFILLHCLS 390
Query: 145 VDSIFQQGWLSFWNPAT---RLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIE 201
L WNP T +++ F F + FGYD STD + VV
Sbjct: 391 --------HLCVWNPTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDDFLVVHACYN 546
Query: 202 KIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSR 257
+A+ +F+L N W G I +FP F Y + G L+G I+W+ R
Sbjct: 547 PKHQANCAEIFSL-RANAWKGIEGI-HFPYTHFRYTNRYNQFGSFLNGAIHWLAFR 708
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 63.9 bits (154), Expect = 1e-10
Identities = 37/123 (30%), Positives = 66/123 (53%)
Frame = +1
Query: 305 VGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLF 364
+GVL CLC ++D + THFV+W MK +G +SW +L+ + Y P + + Y +
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVP---NPENFSYSGPYYI- 171
Query: 365 PLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVS 424
+G+ L++ + + ILY+ R N+ + +E+ + F + YVE+LVS
Sbjct: 172 ---SENGEVLLMFEF-------DLILYNPRDNSFKYPKIESGKGW---FDAEVYVETLVS 312
Query: 425 PLR 427
P++
Sbjct: 313 PMK 321
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 63.2 bits (152), Expect = 2e-10
Identities = 88/350 (25%), Positives = 145/350 (41%), Gaps = 33/350 (9%)
Frame = +3
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLH--LHRSALHGKPHLLLQR 84
L +L++ VLS LP K L KCVCKSW L I+D F+ + ++ S + HLL+ R
Sbjct: 66 LPGELVSNVLSRLPSKVLLLCKCVCKSWFDL-ITDPHFVSNYYVVYNSLQSQEEHLLVIR 242
Query: 85 ENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRY--HVVGCCNGLICLHG 142
+ + ++ LS ++ ++ S NP Y +Y ++G CNG+ L G
Sbjct: 243 RPFFSGLKTY-ISVLSWNTNDPKKHV-SSDVLNPPYEYNSDHKYWTEILGPCNGIYFLEG 416
Query: 143 YSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKN----------FDVKLSFGYDNSTDT 192
NP LM+ LG F K+ F FG+D T+
Sbjct: 417 ---------------NPNV-LMNPSLGEFKALPKSHFTSPHGTYTFTDYAGFGFDPKTND 548
Query: 193 YKVVFFEIEKIRRAS-------LVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVND-G 244
YKVV + ++ +++L+ N WR + LLP + WG +
Sbjct: 549 YKVVVLKDLWLKETDEREIGYWSAELYSLNS----NSWRKLDP-SLLPLPIEIWGSSRVF 713
Query: 245 VHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLD----EVPS 300
+ + +W ++S Q +L+ D+ E++ + PK D + +
Sbjct: 714 TYANNCCHWWGFVEES---------DATQDVVLAFDMVKESFRKIRVPKIRDSSDEKFGT 866
Query: 301 VVP-----SVGVLMNCLCFSHDLEGT--HFVIWQMKKFGDAKSWTQLLKV 343
+VP S+G L+ + + GT F +W MK + D SW + V
Sbjct: 867 LVPFEESASIGFLV------YPVRGTEKRFDVWVMKDYWDEGSWVKQYSV 998
>BI787361
Length = 423
Score = 58.9 bits (141), Expect = 4e-09
Identities = 47/136 (34%), Positives = 68/136 (49%), Gaps = 1/136 (0%)
Frame = +3
Query: 43 SLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGS 102
+L R + V ++WNSLI D TF+KLHL RS + H+LL+ + + V S
Sbjct: 3 ALMRFRYVSETWNSLIF-DPTFVKLHLERSPKN--THVLLEFQAIYDRDVGQQVGVAPCS 173
Query: 103 SLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHG-YSVDSIFQQGWLSFWNPAT 161
LVENP S + L K + G CNGL+C+ + V ++ WNPAT
Sbjct: 174 IRRLVENP--SFTIDDCLTLFKHTN-SIFGSCNGLVCMTKCFDVREFEEECQYRLWNPAT 344
Query: 162 RLMSEKLGCFCVKTKN 177
+MSE C++ K+
Sbjct: 345 GIMSEYSPPLCIQFKD 392
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 55.1 bits (131), Expect = 6e-08
Identities = 47/123 (38%), Positives = 61/123 (49%), Gaps = 2/123 (1%)
Frame = +1
Query: 21 NLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKP-H 79
N T L +LI E+L LPVKSL R KCVCKS+ SL ISD F+ H AL P H
Sbjct: 91 NQSLTTLPQELIREILLRLPVKSLLRFKCVCKSFLSL-ISDPQFVISHY---ALAASPTH 258
Query: 80 LLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKK-GDRYHVVGCCNGLI 138
L+ R + + + S +V P + L P RL G R ++G C GL+
Sbjct: 259 RLILRSHDFYAQSIATESVFKTCSRDVVYFP-LPLPSIPCLRLDDFGIRPKILGSCRGLV 435
Query: 139 CLH 141
L+
Sbjct: 436 LLY 444
>TC235149
Length = 435
Score = 53.9 bits (128), Expect = 1e-07
Identities = 32/105 (30%), Positives = 55/105 (51%)
Frame = +2
Query: 323 FVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLFPLCLHDGDTLILASYSDR 382
FV+W ++FG +SWT+LL V Y+ + H + + PLC+ + + ++L +
Sbjct: 2 FVVWLTREFGVERSWTRLLNVSYEHFRN---HGCPPYYRFVTPLCMSENEDVLLLAND-- 166
Query: 383 YDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVSPLR 427
+G+E + Y+ R N +R ++ Y F S YV SLV P +
Sbjct: 167 -EGSEFVFYNLRDNRIDR--IQDFDSYKFSFLSHDYVPSLVLPYK 292
>AW395660
Length = 381
Score = 49.7 bits (117), Expect = 3e-06
Identities = 25/47 (53%), Positives = 34/47 (72%)
Frame = +3
Query: 20 DNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIK 66
++LP L ++L+ E+LS LPVKSL + +CVCKSW SLI D F+K
Sbjct: 243 ESLPLPFLPDELVVEILSRLPVKSLLQFRCVCKSWMSLIY-DPYFMK 380
>BM143422
Length = 424
Score = 49.3 bits (116), Expect = 3e-06
Identities = 33/62 (53%), Positives = 35/62 (56%)
Frame = +1
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
DLI +L LPVKS+TR KCVCKSW SL ISD F H AL H LL R N
Sbjct: 238 DLIELILLKLPVKSVTRFKCVCKSWLSL-ISDPQFGFSHFD-LALAVPSHRLLLRSNEFS 411
Query: 90 DH 91
H
Sbjct: 412 VH 417
>TC223618
Length = 444
Score = 47.8 bits (112), Expect = 1e-05
Identities = 40/144 (27%), Positives = 61/144 (41%), Gaps = 11/144 (7%)
Frame = +1
Query: 98 HLSGSSLSLVENPWISLAHNPRYRLKKGDRYH-VVGCCNGLICLHGYSVDSIFQQGWLSF 156
HL SLS + N ++ Y +K R+ +VG CNGL+C + +
Sbjct: 28 HLKSCSLSSLFNNLSTVCDELNYPVKNKFRHDGIVGSCNGLLCF-------AIKGDCVLL 186
Query: 157 WNPATRLMSE--------KLGCFCVKTKNFDVKLSFGYDNSTDTYKV--VFFEIEKIRRA 206
WNP+ R+ + + GCF GYD+ + YKV VF + +
Sbjct: 187 WNPSIRVSKKSPPLGNNWRPGCF--------TAFGLGYDHVNEDYKVVAVFCDPSEYFIE 342
Query: 207 SLVSVFTLDGCNGWNGWRDIQNFP 230
V V+++ N WR IQ+FP
Sbjct: 343 CKVKVYSM----ATNSWRKIQDFP 402
>AI443156
Length = 414
Score = 47.8 bits (112), Expect = 1e-05
Identities = 48/153 (31%), Positives = 72/153 (46%)
Frame = +3
Query: 7 NTVTVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIK 66
N +TV +M NLP +++ E+LS LPVKS+ RL+ CK W S II + FI
Sbjct: 60 NPITVSNMA-----NLPV-----EVVTEILSRLPVKSVIRLRSTCKWWRS-IIDSRHFIL 206
Query: 67 LHLHRSALHGKPHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGD 126
HL++S L+L+ + + L SL L NP+ L+H + +
Sbjct: 207 FHLNKS----HTSLILRHRSQLYS--------LDLKSL-LDPNPF-ELSHP---LMCYSN 335
Query: 127 RYHVVGCCNGLICLHGYSVDSIFQQGWLSFWNP 159
V+G NGL+C+ D ++ WNP
Sbjct: 336 SIKVLGSSNGLLCISNVXDD-------IALWNP 413
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 47.8 bits (112), Expect = 1e-05
Identities = 21/46 (45%), Positives = 30/46 (64%)
Frame = +1
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRS 72
L L+ E+L W+ V + +L+CVCK W SL++ D F+K HLH S
Sbjct: 196 LPEGLMIEILVWIRVSNPLQLRCVCKRWKSLVV-DPQFVKKHLHTS 330
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 47.0 bits (110), Expect = 2e-05
Identities = 40/121 (33%), Positives = 57/121 (47%)
Frame = +1
Query: 20 DNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPH 79
D+LPR +++ E+LS LPV+SL R + KSW SLI S Q LHL RS
Sbjct: 34 DHLPR-----EVLTEILSRLPVRSLLRFRSTSKSWKSLIDS-QHLNWLHLTRSLTLASNT 195
Query: 80 LLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLIC 139
L+ R +S F +P +SL H + + ++G CNGL+C
Sbjct: 196 SLILRVDSDLYQTNFPTL-----------DPPVSLNHP---LMCYSNSITLLGSCNGLLC 333
Query: 140 L 140
+
Sbjct: 334 I 336
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 43.9 bits (102), Expect = 1e-04
Identities = 24/46 (52%), Positives = 28/46 (60%)
Frame = +3
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALH 75
+LI E+L PV+S+ R KCVCKSW SL ISD F L S H
Sbjct: 42 ELIREILLRSPVRSVLRFKCVCKSWLSL-ISDPQFTHFDLAASPTH 176
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/49 (51%), Positives = 30/49 (61%)
Frame = -2
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALH 75
L ++I E+L LPVKSL K VCKS SL IS+ F K H R+A H
Sbjct: 540 LPQEIIIEILLRLPVKSLPSFKFVCKS*LSL-ISNPHFAKWHFERNAAH 397
>TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%)
Length = 1228
Score = 41.2 bits (95), Expect = 0.001
Identities = 40/123 (32%), Positives = 53/123 (42%), Gaps = 1/123 (0%)
Frame = +1
Query: 41 VKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHHDHRE-FCVTHL 99
VKSL R K VCK W L + D+ F++ L+ P +L++ +S CV +L
Sbjct: 1 VKSLFRFKTVCKLWYRLSL-DKYFVQ--LYNEVSRKNPMILVEISDSSESKTSLICVDNL 171
Query: 100 SGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIFQQGWLSFWNP 159
G S E +I L DR V CNGL+C SI +G NP
Sbjct: 172 RGVS----EFSFIFL----------NDRVKVRASCNGLLC-----CSSIPDKGVFYVCNP 294
Query: 160 ATR 162
TR
Sbjct: 295 VTR 303
>CD487321
Length = 638
Score = 40.0 bits (92), Expect = 0.002
Identities = 18/34 (52%), Positives = 20/34 (57%)
Frame = +2
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIIS 60
L+ DL+ VLSWLP S RL VCK W S S
Sbjct: 326 LNEDLLERVLSWLPTSSFFRLTSVCKRWKSAAAS 427
>TC211029 similar to UP|Q96VI4 (Q96VI4) Protease 1, partial (3%)
Length = 858
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/56 (44%), Positives = 31/56 (54%), Gaps = 4/56 (7%)
Frame = +3
Query: 25 TQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSA----LHG 76
T L DLI +LS LP+ +L R VCK W+S IIS +F L H + LHG
Sbjct: 153 TNLSTDLIELILSLLPIPTLIRASTVCKLWHS-IISSSSFSTLSNHLNQPWFFLHG 317
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 37.7 bits (86), Expect = 0.010
Identities = 44/199 (22%), Positives = 85/199 (42%), Gaps = 12/199 (6%)
Frame = +3
Query: 154 LSFWNPATRLMSEKLGCFCVKTKNFDVKL------SFGYDNSTDTYKVV----FFEIEKI 203
++FWNP+ R + ++ D L FG+D+ T YK+V F ++
Sbjct: 30 IAFWNPSLRQHRILPYLPVPRRRHPDTTLFAARVCGFGFDHKTRDYKLVRISYFVDLHDR 209
Query: 204 RRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPY 263
+ V ++TL N W+ + P LP++ GV + +++W+ +R K P
Sbjct: 210 SFDAQVKLYTLRA----NAWKTL---PSLPYALC-CARTMGVFVGNSLHWVVTR-KLEPD 362
Query: 264 YNPVTITVEQLGILSLDLRTETYTLLSPPK--GLDEVPSVVPSVGVLMNCLCFSHDLEGT 321
+ I++ DL + + L P G+D + + +L LC + + T
Sbjct: 363 QPDL--------IIAFDLTHDIFRELPLPDTGGVDGGFEI--DLALLGGSLCMTVNFHKT 512
Query: 322 HFVIWQMKKFGDAKSWTQL 340
+W M+++ SW ++
Sbjct: 513 RIDVWVMREYNRRDSWCKV 569
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,177,882
Number of Sequences: 63676
Number of extensions: 464449
Number of successful extensions: 2631
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 2577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2613
length of query: 440
length of database: 12,639,632
effective HSP length: 100
effective length of query: 340
effective length of database: 6,272,032
effective search space: 2132490880
effective search space used: 2132490880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0176a.1


