

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
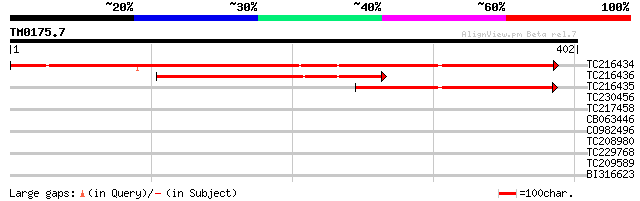
Query= TM0175.7
(402 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216434 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 427 e-120
TC216436 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 180 9e-46
TC216435 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 163 1e-40
TC230456 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, parti... 30 1.9
TC217458 UP|Q39682 (Q39682) Glycine-rich protein (Fragment), par... 29 3.3
CB063446 28 5.7
CO982496 28 9.6
TC208980 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like prot... 28 9.6
TC229768 28 9.6
TC209589 similar to UP|Q9LU28 (Q9LU28) RNA methyltransferase-lik... 28 9.6
BI316623 28 9.6
>TC216434 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (96%)
Length = 1436
Score = 427 bits (1097), Expect = e-120
Identities = 238/392 (60%), Positives = 286/392 (72%), Gaps = 3/392 (0%)
Frame = +1
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDL++SI QT+VALSI+K L SKE ++KNLV+SP SLH +LS++A GS G TLD+L S
Sbjct: 10 MDLRESISNQTDVALSISKLLLSKEA-RDKNLVYSPLSLHVVLSIIAAGSKGPTLDQLLS 186
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FLR S DHLN+F SQ+ + VLSD +P+ LSF + +W ++SL+L SFKQL++ YK
Sbjct: 187 FLRSKSTDHLNSFASQLFAVVLSDASPAGGPRLSFADGVWVEQSLSLLPSFKQLVSADYK 366
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
ATLA VDF+TK +V EVNSW EKETNGL+ +LLP +V T+LIFANALYFKG W
Sbjct: 367 ATLASVDFQTKAVEVANEVNSWAEKETNGLVKDLLPPGSVDSSTRLIFANALYFKGAWNE 546
Query: 179 TFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMC 238
FD S+T DFHLL+G SI VPFM+S +K QFIR FDGF VL L YKQG DK+ +F+M
Sbjct: 547 KFDSSITKDYDFHLLDGRSIRVPFMTS-RKNQFIRAFDGFKVLGLPYKQGEDKR-QFTMY 720
Query: 239 IFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGV 298
FLP KDGL +L EKLASES FL+ KLP K+ V FRIP+FKI F EASNVLKELGV
Sbjct: 721 FFLPETKDGLLALAEKLASESGFLERKLPNNKLEVGDFRIPRFKISFGFEASNVLKELGV 900
Query: 299 VSPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSART 357
V PFS T+MV S + + L V I HK+ IEVNEEGTEA A + C T
Sbjct: 901 VLPFSV--GGLTEMVDSAVGQNLFVSDIFHKSFIEVNEEGTEAAAATAATIQFGCAMFPT 1074
Query: 358 GIDFVADHPFLFLIREDLTGTILFIGQVLHPE 389
IDFVADHPFLFLIREDLTGT+LFIGQVL+P+
Sbjct: 1075EIDFVADHPFLFLIREDLTGTVLFIGQVLNPQ 1170
>TC216436 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (40%)
Length = 489
Score = 180 bits (457), Expect = 9e-46
Identities = 97/163 (59%), Positives = 118/163 (71%)
Frame = +2
Query: 105 LSHSFKQLMTTHYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKL 164
L SFKQL+ YKATLA VDF+TK +V EVNSW EKETNGL+ +LLP +V TKL
Sbjct: 5 LHPSFKQLVRAQYKATLASVDFQTKAVEVTNEVNSWAEKETNGLVKDLLPPGSVDNSTKL 184
Query: 165 IFANALYFKGVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLS 224
IFANAL+ KG W FD S+T D HLLNG+S++VPFM+S+KK QFI + F VL LS
Sbjct: 185 IFANALWCKGAWNEKFDASITKHYDLHLLNGSSVKVPFMTSKKK-QFIMA*NSFKVLGLS 361
Query: 225 YKQGRDKKLRFSMCIFLPNAKDGLPSLIEKLASESCFLKGKLP 267
YKQG DK+ +F+M FLP KDGL +L EKLA +S FL+ +LP
Sbjct: 362 YKQGEDKR-QFTMYFFLPETKDGLLALAEKLAFDSGFLERRLP 487
>TC216435 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (37%)
Length = 820
Score = 163 bits (413), Expect = 1e-40
Identities = 94/144 (65%), Positives = 103/144 (71%), Gaps = 1/144 (0%)
Frame = +2
Query: 246 DGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKS 305
DGL +L EKLASES FL+ KLP QKV V FRIP+FKI F E SNVLKELGVV PFS
Sbjct: 11 DGLLALAEKLASESGFLERKLPNQKVEVGDFRIPRFKISFGFEVSNVLKELGVVLPFSV- 187
Query: 306 NANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFVAD 364
T+MV SP+ + LCV +I HK+ IEVNEEGTEA A S T IDFVAD
Sbjct: 188 -GGLTEMVDSPVGQNLCVSNIFHKSFIEVNEEGTEAAAATSATIRLRSAMLPTKIDFVAD 364
Query: 365 HPFLFLIREDLTGTILFIGQVLHP 388
HPFLFLIREDLTGT+LFIGQVL P
Sbjct: 365 HPFLFLIREDLTGTVLFIGQVLDP 436
>TC230456 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, partial (72%)
Length = 907
Score = 30.0 bits (66), Expect = 1.9
Identities = 19/65 (29%), Positives = 32/65 (49%)
Frame = +3
Query: 101 KSLALSHSFKQLMTTHYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHK 160
+SL L H F ++ +YK TL R V +E+++ + T+L P +AV+
Sbjct: 24 ESLKLEHQFLRVPFEYYKKTL-----RANHRAVEKEMSAVISGVNEAAATDLSPDDAVNH 188
Query: 161 LTKLI 165
L L+
Sbjct: 189 LNSLV 203
>TC217458 UP|Q39682 (Q39682) Glycine-rich protein (Fragment), partial (9%)
Length = 918
Score = 29.3 bits (64), Expect = 3.3
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Frame = -1
Query: 293 LKELGVVSPFSKSNANFTKMVVSPLDELCVE-SIHHK 328
L +GVV P+ +N+NF ++ + +D + S HH+
Sbjct: 678 LNNVGVVPPYRNTNSNFIGLIATDIDSITATLSRHHR 568
>CB063446
Length = 422
Score = 28.5 bits (62), Expect = 5.7
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = -1
Query: 212 IRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSLIEKLASESC 260
+RTFDGFN D + FS C++ +++ PS S++C
Sbjct: 422 LRTFDGFNC-------AIDFVITFSSCVYKQSSRRVRPSAFNACPSKTC 297
>CO982496
Length = 596
Score = 27.7 bits (60), Expect = 9.6
Identities = 27/96 (28%), Positives = 39/96 (40%)
Frame = +1
Query: 44 SVMAVGSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVLSDTTPSFLLSFVNEMWADKSL 103
+V +G+ GST L H +LS +L+ TP+ LLSF N S
Sbjct: 220 NVSVIGNKGSTWIPSLHSLHVAGAHH------SLLSLLLAHRTPTILLSFAN----SSSY 369
Query: 104 ALSHSFKQLMTTHYKATLALVDFRTKGDQVCREVNS 139
LS +F T ++ A + T D V + S
Sbjct: 370 KLSSAF-----TCHQVCAARISNATNVDPVSTRLES 462
>TC208980 similar to UP|Q9FF46 (Q9FF46) ABC transporter-like protein, partial
(15%)
Length = 820
Score = 27.7 bits (60), Expect = 9.6
Identities = 31/141 (21%), Positives = 52/141 (35%), Gaps = 13/141 (9%)
Frame = +1
Query: 49 GSAGSTLDELFSFLRFDSVDHLNT------------FFSQVLSTVLSDTTPSFLLSF-VN 95
G + S + L FL D++DH NT FF+ +ST + L + V
Sbjct: 67 GESDSGMSSLAYFLSKDTIDHXNTLIKPVVYLSMFYFFTNPISTFADNYVVLLCLVYCVT 246
Query: 96 EMWADKSLALSHSFKQLMTTHYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPT 155
+ S+ QL + L L+ + K +V + + + +
Sbjct: 247 GIAYALSIFFEPGAAQLWSVLLPVVLTLIATQPKDSKVLKNIANLCYSKW---------- 396
Query: 156 NAVHKLTKLIFANALYFKGVW 176
L L+ ANA ++GVW
Sbjct: 397 ----ALQALVVANAERYQGVW 447
>TC229768
Length = 1023
Score = 27.7 bits (60), Expect = 9.6
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 2/105 (1%)
Frame = -3
Query: 190 FHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLP 249
FHLL GTS E+ + S + + FDG ++ S G L S C P+ + P
Sbjct: 730 FHLLRGTSSEISAIVSSSSSSGL--FDG-SICDFSISLGLRSALLLS*C---PSESNSCP 569
Query: 250 SLIEKLA--SESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNV 292
+ A S L +LP + + +P + CF S+V
Sbjct: 568 LIFSLKAPLDSSTSLSSELPCGTLSLSSESLP-LQSCFSSSDSSV 437
>TC209589 similar to UP|Q9LU28 (Q9LU28) RNA methyltransferase-like protein,
partial (52%)
Length = 1491
Score = 27.7 bits (60), Expect = 9.6
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = -3
Query: 159 HKLTKLIFANALYFKGVWKH 178
H+L KL AN L +GVW+H
Sbjct: 886 HRLLKLQKANTLNMRGVWEH 827
>BI316623
Length = 408
Score = 27.7 bits (60), Expect = 9.6
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = -1
Query: 159 HKLTKLIFANALYFKGVWKH 178
H+L KL AN L +GVW+H
Sbjct: 270 HRLLKLQKANTLNMRGVWEH 211
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,259,519
Number of Sequences: 63676
Number of extensions: 232172
Number of successful extensions: 1193
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1184
length of query: 402
length of database: 12,639,632
effective HSP length: 99
effective length of query: 303
effective length of database: 6,335,708
effective search space: 1919719524
effective search space used: 1919719524
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0175.7


