

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
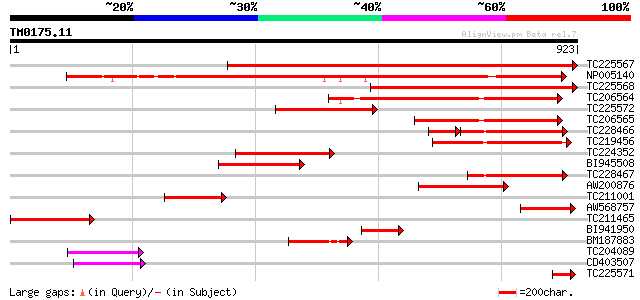
Query= TM0175.11
(923 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 1042 0.0
NP005140 heat shock protein 652 0.0
TC225568 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 628 e-180
TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 377 e-104
TC225572 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 296 4e-80
TC206565 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-l... 262 6e-70
TC228466 similar to UP|ERD1_ARATH (P42762) ERD1 protein, chlorop... 179 1e-65
TC219456 homologue to UP|Q9LLI0 (Q9LLI0) ClpB, partial (24%) 223 3e-58
TC224352 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 193 3e-49
BI945508 homologue to PIR|T51523|T51 clpB heat shock protein-lik... 192 4e-49
TC228467 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, ... 162 5e-40
AW200876 weakly similar to GP|4105131|gb| ClpC protease {Spinaci... 146 5e-35
TC211001 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 140 3e-33
AW568757 similar to SP|P35100|CLPA_ ATP-dependent clp protease A... 140 3e-33
TC211465 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 109 6e-24
BI941950 similar to PIR|T51523|T51 clpB heat shock protein-like ... 87 3e-17
BM187883 homologue to GP|9651530|gb| ClpB {Phaseolus lunatus}, p... 74 3e-13
TC204089 69 1e-11
CD403507 64 3e-10
TC225571 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 60 3e-09
>TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (61%)
Length = 2077
Score = 1042 bits (2694), Expect = 0.0
Identities = 530/569 (93%), Positives = 552/569 (96%)
Frame = +3
Query: 355 KKLMEEIKQNDNIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYR 414
KKLMEEIKQ+D IILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYR
Sbjct: 6 KKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYR 185
Query: 415 KHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQY 474
KHIEKDPALERRFQPV+VPEPTVDE+IQIL+GLRERYE HHKL YTD+ALVAA+QLS+QY
Sbjct: 186 KHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDEALVAAAQLSYQY 365
Query: 475 ISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAGE 534
ISDRFLPDKAIDLIDEAGSRVRL+HAQLPEEARELDKEVRQI+KEK+EAVRNQDFEKAGE
Sbjct: 366 ISDRFLPDKAIDLIDEAGSRVRLQHAQLPEEARELDKEVRQIIKEKEEAVRNQDFEKAGE 545
Query: 535 LRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSDE 594
LRDREMDLK QIS L+EKGKEMSKAE+EA GP+VTE DIQHIV+SWTG+PV+KVS+DE
Sbjct: 546 LRDREMDLKAQISTLVEKGKEMSKAETEAGDEGPIVTEADIQHIVSSWTGIPVEKVSTDE 725
Query: 595 SDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELA 654
SDRLLKMEETLHKRVIGQDEAVKAI RAIRRARVGLKNPNRPIASFIFSGPTGVGKSELA
Sbjct: 726 SDRLLKMEETLHKRVIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELA 905
Query: 655 KTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVL 714
K LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVL
Sbjct: 906 KALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVL 1085
Query: 715 FDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFD 774
FDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFD
Sbjct: 1086FDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFD 1265
Query: 775 LDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFD 834
LDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFD
Sbjct: 1266LDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFD 1445
Query: 835 RLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSV 894
RLK K+I+L VTERFRDRVVEEGY+PSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSV
Sbjct: 1446RLKVKDIELQVTERFRDRVVEEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSV 1625
Query: 895 IIDADSDGKVIVLNGSSGAPESLPEALPV 923
I+D DSDG VIVLNGSSGAPESLPE LPV
Sbjct: 1626IVDVDSDGNVIVLNGSSGAPESLPETLPV 1712
>NP005140 heat shock protein
Length = 2736
Score = 652 bits (1681), Expect = 0.0
Identities = 367/876 (41%), Positives = 534/876 (60%), Gaps = 62/876 (7%)
Frame = +1
Query: 93 ERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVE 152
E+FT K + + + E A GH + + L+ + GI + + G + AR
Sbjct: 10 EKFTHKTNEALASAHELAMSSGHAQLTPIHLAHALISDPNGIFVLAINSAGGGEESARA- 186
Query: 153 VEKTIGRGSGFVAV------EIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREG 206
VE+ + + + E+P + R + + + G + + L+LG+L +
Sbjct: 187 VERVLNQALKKLPCQSPPPDEVPASTNLVRAIRRAQAAQKSRGDTRLAVDQLILGILEDS 366
Query: 207 EGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKL 266
+ +L++ G + ++V ++ G+ G V L+ YG +L +
Sbjct: 367 Q--IGDLLKEAGVAVAKVESEVDKLRGKE-----GKKVESASGDTNFQALKTYGRDLVEQ 525
Query: 267 ANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPET 326
A GKLDPV+GR ++I RV++IL RRTKNNP LVGEPGVGKTA+ EGLAQRI GDVP
Sbjct: 526 A--GKLDPVIGRDEEIRRVVRILSRRTKNNPVLVGEPGVGKTAVVEGLAQRIVRGDVPSN 699
Query: 327 IEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDN-IILFIDEVHTLIGAGAAEG 385
+ +I LDMG LVAG KYRGEFEERLK +++E+++ + +ILFIDE+H ++GAG EG
Sbjct: 700 LADVRLIALDMGALVAGAKYRGEFEERLKAVLKEVEEAEGKVILFIDEIHLVLGAGRTEG 879
Query: 386 AIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILR 445
++DAAN+ KP LARG+L+CIGATTL+EYRK++EKD A ERRFQ V V EP+V ++I ILR
Sbjct: 880 SMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVFVAEPSVVDTISILR 1059
Query: 446 GLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEE 505
GL+ERYE HH + D ALV A+QLS++YI+ R LPDKAIDL+DEA + VR++ PEE
Sbjct: 1060GLKERYEGHHGVRIQDRALVMAAQLSNRYITGRHLPDKAIDLVDEACANVRVQLDSQPEE 1239
Query: 506 ARELDK-------EVRQIVKEKDEAVRNQDFEKAGELRD-------------REMDLKTQ 545
L++ E+ + KEKD+A + + E EL D +E + +
Sbjct: 1240IDNLERKRMQLEVELHALEKEKDKASKARLVEVRKELDDLRDKLQPLMMKYRKEKERVDE 1419
Query: 546 ISALIEKGKEMSKAESEADGAGPVVTEVDIQH---------------------------- 577
I L +K +E+ A EA+ + D+++
Sbjct: 1420IRRLKKKREELLFALQEAERRYDLARAADLRYGAIQEVETAIQQLEGSTEENLMLTETVG 1599
Query: 578 ------IVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLK 631
+V+ WTG+PV ++ +E +RL+ + + LH RV+GQD+AV A+ A+ R+R GL
Sbjct: 1600PEQIAEVVSRWTGIPVTRLGQNEKERLIGLGDRLHSRVVGQDQAVNAVAEAVLRSRAGLG 1779
Query: 632 NPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPG 691
P +P SF+F GPTGVGK+ELAK LA F +E ++R+DMSE+ME+H+VS+LIG+PPG
Sbjct: 1780RPQQPTGSFLFLGPTGVGKTELAKALAEQLFDNENQLVRIDMSEYMEQHSVSRLIGAPPG 1959
Query: 692 YVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNT 751
YVG+ EGGQLTEAVRRRPY+VVLFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+NT
Sbjct: 1960YVGHEEGGQLTEAVRRRPYSVVLFDEVEKAHTSVFNTLLQVLDDGRLTDGQGRTVDFRNT 2139
Query: 752 LLIMTSNVGSSVIEKG-GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEM 810
++IMTSN+G+ + G K + D V +E+++ FRPE LNRLDE+
Sbjct: 2140VIIMTSNLGAEHLLSGLSGKCTMQVARDR-----------VMQEVRRQFRPELLNRLDEI 2286
Query: 811 IVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRA 870
+VF L+ +++++A + +K+V RL K I L+VT+ D ++ E YDP YGARP+RR
Sbjct: 2287VVFDPLSHDQLRKVARLQMKDVASRLAEKGIALAVTDAALDYILSESYDPVYGARPIRRW 2466
Query: 871 IMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIV 906
+ + + ++ ++ EI E +V IDA +G +V
Sbjct: 2467LEKKVVTELSRMLVREEIDENSTVYIDAGPNGGELV 2574
>TC225568 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (36%)
Length = 1287
Score = 628 bits (1619), Expect = e-180
Identities = 322/336 (95%), Positives = 330/336 (97%)
Frame = +2
Query: 588 DKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTG 647
+KVS+DESDRLLKMEETLHKRVIGQDEAVKAI RAIRRARVGLKNPNRPIASFIFSGPTG
Sbjct: 5 EKVSTDESDRLLKMEETLHKRVIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTG 184
Query: 648 VGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRR 707
VGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRR
Sbjct: 185 VGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRR 364
Query: 708 RPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 767
RPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG
Sbjct: 365 RPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 544
Query: 768 GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADI 827
GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADI
Sbjct: 545 GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADI 724
Query: 828 MLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGE 887
MLKEVFDRLK KEIDLSVTERFR+RVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA E
Sbjct: 725 MLKEVFDRLKAKEIDLSVTERFRERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLARE 904
Query: 888 IKEGDSVIIDADSDGKVIVLNGSSGAPESLPEALPV 923
IKEGDSVI+DADSDG VIVLNGSSGAP+SL EALPV
Sbjct: 905 IKEGDSVIVDADSDGNVIVLNGSSGAPDSLEEALPV 1012
>TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (40%)
Length = 1497
Score = 377 bits (968), Expect = e-104
Identities = 194/389 (49%), Positives = 267/389 (67%), Gaps = 8/389 (2%)
Frame = +2
Query: 519 EKDEAVRNQDFEKAGELR-------DREMD-LKTQISALIEKGKEMSKAESEADGAGPVV 570
E +A R D +A EL+ R+++ + ++ + GK M + E V
Sbjct: 8 EIQQAEREYDLNRAAELKYGSLNSLQRQLESAEKELDEYMNSGKSMLREE---------V 160
Query: 571 TEVDIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGL 630
T DI IV+ WTG+PV K+ E ++LL +EE LHKRV+GQD AVKAI AI+R+R GL
Sbjct: 161 TGNDIAEIVSKWTGIPVSKLQQSEREKLLHLEEVLHKRVVGQDPAVKAIAEAIQRSRAGL 340
Query: 631 KNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPP 690
+P+RPIASF+F GPTGVGK+ELAK LA+Y F +EEA++R+DMSE+ME+H VS+LIG+PP
Sbjct: 341 SDPHRPIASFMFMGPTGVGKTELAKALAAYLFNTEEALVRIDMSEYMEKHAVSRLIGAPP 520
Query: 691 GYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKN 750
GYVGY EGGQLTE VRRRPY V+LFDEIEKAH DVFN+ LQIL+DGR+TDS+GRTV F N
Sbjct: 521 GYVGYEEGGQLTEIVRRRPYAVILFDEIEKAHADVFNVFLQILDDGRVTDSQGRTVSFTN 700
Query: 751 TLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEM 810
T++IMTSNVGS I + D D K+ +Y IK V + + FRPEF+NR+DE
Sbjct: 701 TVIIMTSNVGSQYI------LNTDDDTTPKELAYETIKQRVMDAARSIFRPEFMNRVDEY 862
Query: 811 IVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRA 870
IVF+ L + ++ I + L+ V R+ +++ + VT+ + GYDP+YGARP++R
Sbjct: 863 IVFQPLDREQISSIVRLQLERVQKRIADRKMKIQVTDAAVQLLGSLGYDPNYGARPVKRV 1042
Query: 871 IMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
I + +E+ +A+ +L GE KE D+++ID +
Sbjct: 1043IQQNVENELAKGILRGEFKEEDAILIDTE 1129
>TC225572 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (18%)
Length = 511
Score = 296 bits (757), Expect = 4e-80
Identities = 148/167 (88%), Positives = 161/167 (95%)
Frame = +3
Query: 433 PEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAG 492
PEPTV+E+IQIL+GLRERYE HHKL YTDDALVAA+QLSHQYISDRFLPDKAIDLIDEAG
Sbjct: 9 PEPTVNETIQILKGLRERYEIHHKLHYTDDALVAAAQLSHQYISDRFLPDKAIDLIDEAG 188
Query: 493 SRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAGELRDREMDLKTQISALIEK 552
SRVRL+HAQLPEEARELDKEVRQIVKEK+E+VRNQDFEKAGELRD+EMDLK QISALIEK
Sbjct: 189 SRVRLQHAQLPEEARELDKEVRQIVKEKEESVRNQDFEKAGELRDKEMDLKAQISALIEK 368
Query: 553 GKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSDESDRLL 599
GKEMSKAESEA GP+VTEVDIQHIV+SWTG+PV+KVS+DESDRLL
Sbjct: 369 GKEMSKAESEAGDEGPMVTEVDIQHIVSSWTGIPVEKVSTDESDRLL 509
>TC206565 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(25%)
Length = 1078
Score = 262 bits (669), Expect = 6e-70
Identities = 128/241 (53%), Positives = 176/241 (72%)
Frame = +2
Query: 659 SYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEI 718
SY F +EEA++R+DMSE+ME+HTVS+LIG+PPGYVGY EGGQLTE VRRRPY V+LFDEI
Sbjct: 2 SYLFNTEEALVRIDMSEYMEKHTVSRLIGAPPGYVGYEEGGQLTETVRRRPYAVILFDEI 181
Query: 719 EKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYD 778
EKAH DVFN+ LQIL+DGR+TDS+GRTV F NT++IMTSNVGS I + D D
Sbjct: 182 EKAHSDVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSNVGSQYI------LNTDDDTV 343
Query: 779 EKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKT 838
K+S+Y IK V + + FRPEF+NR+DE IVF+ L + ++ I + L+ V R+
Sbjct: 344 PKESAYETIKQRVMDAARSIFRPEFMNRVDEYIVFQPLDRNQISSIVRLQLERVQKRIAD 523
Query: 839 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 898
+++ + VTE + GYDP+YGARP++R I + +E+ +A+ +L GE KE D++++D
Sbjct: 524 RKMKIQVTEAAIQLLGSLGYDPNYGARPVKRVIQQNVENELAKGILRGEFKEEDTILVDT 703
Query: 899 D 899
+
Sbjct: 704 E 706
>TC228466 similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (23%)
Length = 936
Score = 179 bits (454), Expect(2) = 1e-65
Identities = 90/176 (51%), Positives = 131/176 (74%), Gaps = 1/176 (0%)
Frame = +2
Query: 734 EDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRK-IGFDLDYDEKDSSYNRIKSLVT 792
+DG+LTDS+GR V FKN L++MTSNVGSS I KG IGF L D+K +SYN +KS+V
Sbjct: 155 QDGQLTDSQGRRVSFKNALVVMTSNVGSSAIAKGRHNSIGF-LIPDDKTTSYNGLKSMVI 331
Query: 793 EELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDR 852
EEL+ YFRPE LNR+DE++VF+ L K ++ +I D++L+++ R+ + + + V+E ++
Sbjct: 332 EELRSYFRPELLNRIDEVVVFQPLEKSQLLQILDLLLQDMKKRVLSLGVHVKVSEAVKNL 511
Query: 853 VVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 908
V ++GY+P+YGARPLRRAI L+ED ++E L GE K+GD+V+ID D++G V N
Sbjct: 512 VCQQGYNPTYGARPLRRAITSLIEDPLSEAFLYGECKQGDTVLIDLDANGNPFVTN 679
Score = 90.5 bits (223), Expect(2) = 1e-65
Identities = 40/51 (78%), Positives = 48/51 (93%)
Frame = +1
Query: 683 SKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQIL 733
SKLIGSPPGYVGY EGG LTEA+RR+P+T++L DEIEKAHPD+FN++LQIL
Sbjct: 1 SKLIGSPPGYVGYGEGGVLTEAIRRKPFTLLLLDEIEKAHPDIFNILLQIL 153
>TC219456 homologue to UP|Q9LLI0 (Q9LLI0) ClpB, partial (24%)
Length = 871
Score = 223 bits (568), Expect = 3e-58
Identities = 114/227 (50%), Positives = 161/227 (70%)
Frame = +1
Query: 688 SPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVD 747
+PPGYVGY EGGQLTE VRRRPY+VVLFDEIEKAH DVFN++LQ+L+DGR+TDS+GRTV
Sbjct: 1 APPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHHDVFNILLQLLDDGRITDSQGRTVS 180
Query: 748 FKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRL 807
F N ++IMTSN+GS I R D+K + Y+++K V E +Q F PEF+NR+
Sbjct: 181 FTNCVVIMTSNIGSHYILDTLRS-----TQDDKTAVYDQMKRQVVELARQTFHPEFMNRI 345
Query: 808 DEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPL 867
DE IVF+ L ++ +I ++ ++ V +RLK K+IDL TE+ + G+DP++GARP+
Sbjct: 346 DEYIVFQPLDSEQISKIVELQMERVKNRLKQKKIDLHYTEKAVKLLGVLGFDPNFGARPV 525
Query: 868 RRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAP 914
+R I +L+E+ +A +L G+ KE DS+I+DAD + L+G +P
Sbjct: 526 KRVIQQLVENEIAMGVLRGDFKEEDSIIVDAD-----VTLSGKERSP 651
>TC224352 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (17%)
Length = 505
Score = 193 bits (491), Expect = 3e-49
Identities = 94/161 (58%), Positives = 125/161 (77%)
Frame = +1
Query: 368 ILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRF 427
ILFIDE+HT++GAGA+ GA+DA N+LKP L RGEL+CIGATTLDEYRK+IEKDPALERRF
Sbjct: 10 ILFIDEIHTVVGAGASNGAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALERRF 189
Query: 428 QPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDL 487
Q V V +P+V+++I ILRGLRERYE HH + +D ALV A+ LS +YIS RFLPDKAIDL
Sbjct: 190 QQVYVDQPSVEDTISILRGLRERYELHHGVRISDSALVDAAILSDRYISGRFLPDKAIDL 369
Query: 488 IDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQD 528
+DEA +++++ P E+++ V ++ E+ + + D
Sbjct: 370 VDEAAAKLKMEITSKPTALDEINRSVLKLEMERLSLMNDTD 492
>BI945508 homologue to PIR|T51523|T51 clpB heat shock protein-like -
Arabidopsis thaliana, partial (14%)
Length = 432
Score = 192 bits (489), Expect = 4e-49
Identities = 94/141 (66%), Positives = 117/141 (82%), Gaps = 1/141 (0%)
Frame = +1
Query: 340 LVAGTKYRGEFEERLKKLMEEIKQNDN-IILFIDEVHTLIGAGAAEGAIDAANILKPALA 398
L+AG KYRGEFE+RLK +++E+ ++D ILFIDE+HT++GAGA GA+DA N+LKP L
Sbjct: 10 LIAGAKYRGEFEDRLKAVLKEVTESDGQTILFIDEIHTVVGAGATNGAMDAGNLLKPMLG 189
Query: 399 RGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLS 458
RGEL+CIGATTLDEYRK+IEKDPALERRFQ V V +PTV+++I ILRGLRERYE HH +
Sbjct: 190 RGELRCIGATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVR 369
Query: 459 YTDDALVAASQLSHQYISDRF 479
+D ALV A+ LS +YIS RF
Sbjct: 370 ISDSALVEAAILSDRYISGRF 432
>TC228467 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (17%)
Length = 799
Score = 162 bits (411), Expect = 5e-40
Identities = 82/164 (50%), Positives = 121/164 (73%), Gaps = 1/164 (0%)
Frame = +1
Query: 746 VDFKNTLLIMTSNVGSSVIEKGGRK-IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFL 804
V FKN L++MTSNVGSS I KG IGF L D+K +SYN +KS+V EEL+ YFRPE L
Sbjct: 1 VSFKNALVVMTSNVGSSAITKGRHNSIGF-LIPDDKKTSYNGLKSMVIEELRTYFRPELL 177
Query: 805 NRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGA 864
NR+DE++VF+ L K ++ +I D++L+++ R+ + I + V+E ++ V ++GY+P+YGA
Sbjct: 178 NRIDEVVVFQPLEKSQLLQILDVLLQDMKKRVLSLGIHVKVSEAVKNLVCQQGYNPTYGA 357
Query: 865 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 908
RPLRRAI L+ED ++E +L GE K+GD+V++D D++G V N
Sbjct: 358 RPLRRAITSLIEDPLSEALLYGECKQGDTVLVDLDANGNPFVTN 489
>AW200876 weakly similar to GP|4105131|gb| ClpC protease {Spinacia oleracea},
partial (12%)
Length = 487
Score = 146 bits (368), Expect = 5e-35
Identities = 85/147 (57%), Positives = 99/147 (66%)
Frame = +2
Query: 666 EAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDV 725
E +I LDM M+ H+ +LI +P GY GG L EAVRR YTV +I+ AHP V
Sbjct: 47 EGIILLDMCGDMDGHSGLQLIDTPTGYSASYVGG*LLEAVRRHTYTV*RICDIDNAHPAV 226
Query: 726 FNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYN 785
FNM LQILE+GRL+DS + DF NTLLIMTSN G V EK GR I FDLDYDEKDS
Sbjct: 227 FNMKLQILEEGRLSDSLR*SEDFTNTLLIMTSNAGGRVHEKRGRII*FDLDYDEKDSMLY 406
Query: 786 RIKSLVTEELKQYFRPEFLNRLDEMIV 812
RI S VT+ELK + PE LNR ++MIV
Sbjct: 407 RIMS*VTDELKPHCMPESLNRENDMIV 487
>TC211001 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (16%)
Length = 456
Score = 140 bits (353), Expect = 3e-33
Identities = 66/101 (65%), Positives = 80/101 (78%)
Frame = +2
Query: 252 KMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIA 311
K LE+YG +LT +A GKLDPV+GR +I R IQIL RRTKNNP L+GEPGVGKTAI+
Sbjct: 152 KYEALEKYGKDLTAMAKAGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGVGKTAIS 331
Query: 312 EGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEE 352
EGLAQRI GDVP+ + + +I+LDMG L+AG KYRGEFE+
Sbjct: 332 EGLAQRIVQGDVPQALMNRRLISLDMGALIAGAKYRGEFED 454
>AW568757 similar to SP|P35100|CLPA_ ATP-dependent clp protease ATP-binding
subunit clpA homolog chloroplast precursor. [Garden
pea], partial (9%)
Length = 271
Score = 140 bits (352), Expect = 3e-33
Identities = 71/90 (78%), Positives = 80/90 (88%)
Frame = +2
Query: 832 VFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEG 891
VF RLK KEIDLSVTERFR+RVV+EGY+PSYGARPL RAIM+LLEDSMAEKMLA EI EG
Sbjct: 2 VFQRLKAKEIDLSVTERFRERVVDEGYNPSYGARPLTRAIMQLLEDSMAEKMLAREI*EG 181
Query: 892 DSVIIDADSDGKVIVLNGSSGAPESLPEAL 921
DSVI+D+DS+G VIVLNG GAP+SL + L
Sbjct: 182 DSVIVDSDSEGNVIVLNGMFGAPDSLEDVL 271
>TC211465 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (15%)
Length = 593
Score = 109 bits (272), Expect = 6e-24
Identities = 65/139 (46%), Positives = 85/139 (60%), Gaps = 1/139 (0%)
Frame = +2
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT 60
M+RVLAQS+ +PGLV HG S +RS KMMS L+ LRMSGFSGLRT+N LDT
Sbjct: 176 MARVLAQSVNVPGLVAEHRHGQQKGSGKLKRSTKMMSALRTNGLRMSGFSGLRTFNPLDT 355
Query: 61 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFT-EKAIKVIMLSQEEARRLGHNFVG 119
MLRPG+DF SKV T+SR+ARA+RC+ ++ F E + + + L +
Sbjct: 356 MLRPGIDFHSKVSIATSSRQARATRCVAQSHV*AFH*ESNQS*LC*PRRKQDVLVTILLA 535
Query: 120 TEQILLGLVGEGTGIAARV 138
+ L +GTGIAA+V
Sbjct: 536 QNKFYWVLSAKGTGIAAKV 592
Score = 40.8 bits (94), Expect = 0.003
Identities = 16/33 (48%), Positives = 27/33 (81%)
Frame = +1
Query: 173 RAKRVLELSLEEARQLGHNYIGSEHLLLGLLRE 205
+ +++ L+ EEAR+LGHN++G+E +LLGL+ E
Sbjct: 469 KQSKLIMLAQEEARRLGHNFVGTEQILLGLIGE 567
>BI941950 similar to PIR|T51523|T51 clpB heat shock protein-like -
Arabidopsis thaliana, partial (13%)
Length = 421
Score = 87.4 bits (215), Expect = 3e-17
Identities = 38/68 (55%), Positives = 53/68 (77%)
Frame = +2
Query: 574 DIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNP 633
DI IV+ WTG+P+ K+ + ++LL +EE LHKRV+GQD AVKA+ AI+R+R GL +P
Sbjct: 215 DIADIVSKWTGIPISKLQQSDREKLLYLEEELHKRVVGQDPAVKAVAEAIQRSRAGLSDP 394
Query: 634 NRPIASFI 641
+RPIASF+
Sbjct: 395 HRPIASFM 418
>BM187883 homologue to GP|9651530|gb| ClpB {Phaseolus lunatus}, partial (14%)
Length = 427
Score = 73.9 bits (180), Expect = 3e-13
Identities = 40/105 (38%), Positives = 71/105 (67%)
Frame = +3
Query: 454 HHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEV 513
HH + +D ALV+A+ L+ +YI++RFLPDKAIDL+DEA +++++ P E E+D+ +
Sbjct: 3 HHGVKISDSALVSAAVLADRYITERFLPDKAIDLVDEAAAKLKMEITSKPTELDEIDRAI 182
Query: 514 RQIVKEKDEAVRNQDFEKAGELRDREMDLKTQISALIEKGKEMSK 558
++ EK +++N D +KA ++R L+ +S L +K KE+++
Sbjct: 183 LKLEMEK-LSLKN-DTDKAS--KERLSKLENDLSLLKQKQKELTE 305
>TC204089
Length = 1141
Score = 68.6 bits (166), Expect = 1e-11
Identities = 43/128 (33%), Positives = 71/128 (54%), Gaps = 3/128 (2%)
Frame = +2
Query: 94 RFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEV 153
+++E+AIK + + EAR+L + GTE +L+G++ EGT AA+ +A GI+L R E
Sbjct: 362 KWSERAIKSYAMGELEARKLKYPNTGTEALLMGILVEGTSKAAKFSRANGITLLKVREET 541
Query: 154 EKTIGRGS--GFVAVEIPFTPRAKRVLELSLEEARQLGH-NYIGSEHLLLGLLREGEGVA 210
+G+ F P T A++ L+ ++EE + G I + HLLLG+ + E
Sbjct: 542 VGLLGKSDLFFFSPEHPPLTEPAQKALDWAIEEKLKSGEGGEINATHLLLGIWSQKESAG 721
Query: 211 ARVLQDLG 218
++L LG
Sbjct: 722 QQILATLG 745
>CD403507
Length = 633
Score = 63.9 bits (154), Expect = 3e-10
Identities = 43/120 (35%), Positives = 66/120 (54%), Gaps = 4/120 (3%)
Frame = -2
Query: 105 LSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGS--G 162
+ + EAR+L + GTE IL+G++ EGT AA+ L+A GI+L AR E + +G+
Sbjct: 632 MGELEARKLKYPNTGTEAILMGILVEGTSNAAKFLRANGITLLKAREETVELLGKSDLFF 453
Query: 163 FVAVEIPFTPRAKRVLELSLEE-ARQLGH-NYIGSEHLLLGLLREGEGVAARVLQDLGAD 220
F P T A++ L+ ++EE + G I HLLLG+ + E ++L LG D
Sbjct: 452 FSPEHPPLTEPAQKALDWAIEEKLKSAGEGGEINVTHLLLGIWSQEESAGQQILATLGFD 273
>TC225571 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (4%)
Length = 654
Score = 60.5 bits (145), Expect = 3e-09
Identities = 29/38 (76%), Positives = 34/38 (89%)
Frame = +3
Query: 884 LAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESLPEAL 921
LA EIKEGDSVI+D+DS+G VIVLNGSSGAP+SL + L
Sbjct: 3 LAREIKEGDSVIVDSDSEGNVIVLNGSSGAPDSLEDVL 116
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,508,077
Number of Sequences: 63676
Number of extensions: 293655
Number of successful extensions: 1595
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 1568
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1583
length of query: 923
length of database: 12,639,632
effective HSP length: 106
effective length of query: 817
effective length of database: 5,889,976
effective search space: 4812110392
effective search space used: 4812110392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0175.11


