

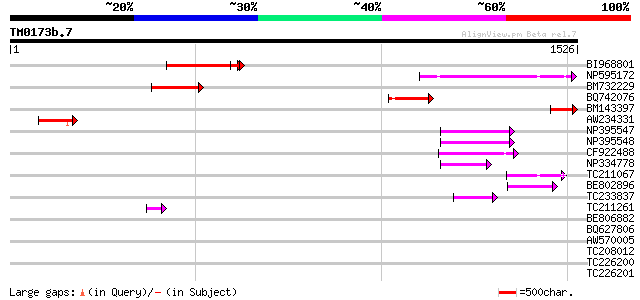
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.7
(1526 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata},... 259 6e-69
NP595172 polyprotein [Glycine max] 226 8e-59
BM732229 191 3e-48
BQ742076 134 2e-31
BM143397 120 3e-27
AW234331 112 2e-24
NP395547 reverse transcriptase [Glycine max] 91 4e-18
NP395548 reverse transcriptase [Glycine max] 80 7e-15
CF922488 76 1e-13
NP334778 reverse transcriptase [Glycine max] 67 6e-11
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 59 1e-08
BE802896 58 3e-08
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 58 4e-08
TC211261 47 5e-05
BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, pa... 43 0.001
BQ627806 42 0.003
AW570005 41 0.005
TC208012 similar to UP|Q761Z7 (Q761Z7) BRI1-KD interacting prote... 40 0.008
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 39 0.013
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 39 0.013
>BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata}, partial
(0%)
Length = 660
Score = 259 bits (662), Expect = 6e-69
Identities = 128/206 (62%), Positives = 160/206 (77%), Gaps = 1/206 (0%)
Frame = -1
Query: 421 NMTMVATAYETSNNCPESLIVEILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNP 480
+MTMVATAY+TS+ C I++ILVAGF GQLK WWDNYLT +EK +I +AVK+D G
Sbjct: 657 HMTMVATAYQTSHECSXXXIIDILVAGFSGQLKRWWDNYLTNEEK*KIYSAVKTDLNGKV 478
Query: 481 II-EDGKFIFDAVNSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTR 539
I +D K I DAVN+LIFTIAQHF+GDPSL KDRS +LLSNLKC++L DFRWY+DTFLTR
Sbjct: 477 ITNDDDKEILDAVNTLIFTIAQHFIGDPSLWKDRSAELLSNLKCRTLADFRWYRDTFLTR 298
Query: 540 VYTREDSQHAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLIAIIQRVAL 599
VYTREDSQ F KEKFLAGLP+S GDKVR+K+RSQ+ G+IPY++LSYGQLI+ +Q+VA
Sbjct: 297 VYTREDSQQPF*KEKFLAGLPRSLGDKVRDKIRSQSANGDIPYESLSYGQLISYVQKVAX 118
Query: 600 KICQDDKIQQQLTKEKSQNRRDLGTF 625
K + + +++ K N+ F
Sbjct: 117 KNLSG*QNSKAISQRKGSNKEGFRFF 40
Score = 58.2 bits (139), Expect = 3e-08
Identities = 25/36 (69%), Positives = 31/36 (85%)
Frame = -2
Query: 595 QRVALKICQDDKIQQQLTKEKSQNRRDLGTFCEQFG 630
+R KICQDDKIQ+QL KEK+Q ++DLG+FCEQFG
Sbjct: 131 KR*XXKICQDDKIQRQLAKEKAQTKKDLGSFCEQFG 24
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 226 bits (575), Expect = 8e-59
Identities = 144/423 (34%), Positives = 218/423 (51%), Gaps = 1/423 (0%)
Frame = +1
Query: 1104 PTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVIN 1163
P K RP + ++ I ++L + +I+ S SP+S V K+ G+ R +
Sbjct: 1789 PVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKD----GSWRFCTD 1956
Query: 1164 YKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFG 1223
Y+ LN +P+P +LL LH A+ FSK D++SG+ QI +Q +DR KTAF G
Sbjct: 1957 YRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHG 2136
Query: 1224 QYEWNVMPFGLKNAPSEFQRIMNEIFN-P*SKFAIVYIDDVLIFSQSIDQHFKHLNTFIS 1282
YEW VMPFGL NAP+ FQ +MN+IF KF +V+ DD+LI+S S H KHL + +
Sbjct: 2137 HYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQ 2316
Query: 1283 IIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFL 1342
+K++ + +K S T++ +LGH + + N ++ +P K QL+ FL
Sbjct: 2317 TLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVK-QLRGFL 2493
Query: 1343 GCLNYVADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAF 1402
G Y F + I PL D L+KD W++ ++K + P L LP+
Sbjct: 2494 GLTGYYRRFIKSYANIAGPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQP 2673
Query: 1403 KIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQS 1462
I+ETDAS IG G +L Q IA+ SK P Q S +E+LAI ++S F+
Sbjct: 2674 FILETDASGIGVGAVLGQ----NGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRH 2841
Query: 1463 DLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDF 1522
L+ KF++R D +S K ++ + ++ + W +DF+IEY G N D
Sbjct: 2842 YLLGNKFIIRTDQRSLKSLMDQSLQTPEQQ----AWLHKFLGYDFKIEYKPGKDNQAADA 3009
Query: 1523 LTR 1525
L+R
Sbjct: 3010 LSR 3018
>BM732229
Length = 421
Score = 191 bits (484), Expect = 3e-48
Identities = 94/139 (67%), Positives = 112/139 (79%), Gaps = 1/139 (0%)
Frame = -3
Query: 383 DLLLEEKDSSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYETSNNCPESLIVE 442
DLLLEE+ +NFKSFSANN+YEWNID + EY I LQ+MTMVATAY+TS+ C E I++
Sbjct: 419 DLLLEERGENNFKSFSANNIYEWNIDAQTEYNIMNTLQHMTMVATAYQTSHECSEETIID 240
Query: 443 ILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNPII-EDGKFIFDAVNSLIFTIAQ 501
ILVAGF GQLKGWWDNYLT +EK++I +AVK+D G I +D K I DAVN+LIFTIAQ
Sbjct: 239 ILVAGFSGQLKGWWDNYLTNEEKSKIYSAVKTDLNGKVITNDDDKEIPDAVNTLIFTIAQ 60
Query: 502 HFVGDPSLIKDRSGDLLSN 520
HF+GDPSL DRS +LLSN
Sbjct: 59 HFIGDPSLW*DRSAELLSN 3
>BQ742076
Length = 429
Score = 134 bits (338), Expect = 2e-31
Identities = 66/120 (55%), Positives = 90/120 (75%)
Frame = -2
Query: 1020 SKPPFVKTLNIISYKEKQINFLKEEISYKNIEVQLQQPSVKSRIENILENIQSSICFDLP 1079
SK P K +N QINFLK+E+S+ NI++QL++P +K +I ++L++IQS+ICFDL
Sbjct: 344 SKLPSKKKIN-------QINFLKDEVSFSNIQIQLEKPQMKEKISSLLKHIQSTICFDLA 186
Query: 1080 NAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSP 1139
AF RK H++ LP +KDF +K IPTK+RPI MNEELLQ+ Q+EI DL K LIR+SK+P
Sbjct: 185 RAF*NRKKHILNLPCDKDFKEKHIPTKSRPI*MNEELLQYCQKEIKDLFNKGLIRKSKNP 6
>BM143397
Length = 408
Score = 120 bits (302), Expect = 3e-27
Identities = 58/70 (82%), Positives = 64/70 (90%)
Frame = -3
Query: 1457 ISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGST 1516
IS FQSDL+NQK L+RVDCK AKDILQKDVKNLASK IFARWQAILSVFDF+IEY K ++
Sbjct: 406 ISKFQSDLLNQKILIRVDCKLAKDILQKDVKNLASKQIFARWQAILSVFDFDIEYFKDTS 227
Query: 1517 NSLPDFLTRE 1526
NSLPD+LTRE
Sbjct: 226 NSLPDYLTRE 197
>AW234331
Length = 370
Score = 112 bits (279), Expect = 2e-24
Identities = 52/112 (46%), Positives = 72/112 (63%), Gaps = 9/112 (8%)
Frame = +2
Query: 79 FVIDKEGIRADFYSDRWTKQREWFFANYQGKRRKKVQDRFYGFLELVQQNIFFFDWFHAY 138
F IDKE +R DFYS QR WFF +Y+G RK+++D+FY F+E V+ NI FFDWFHAY
Sbjct: 11 FSIDKETLRKDFYSSENEPQRRWFFQHYKGTNRKQIKDKFYKFVERVKINILFFDWFHAY 190
Query: 139 TIKKKIPYPYQVDVI---------SWQLKDGKETLSNLPPKTPFFLKGAQSA 181
I+K I YP++ D+I +W++KDG+ S LPP T + L + +
Sbjct: 191 AIRKDIDYPWKQDIIGDPTTNVITNWKIKDGELIQSELPPATQYQLPNVKDS 346
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 90.9 bits (224), Expect = 4e-18
Identities = 60/201 (29%), Positives = 96/201 (46%), Gaps = 1/201 (0%)
Frame = +1
Query: 1159 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1218
R+ I+Y+ LN+A YP+P +L RL + D SG+ QI + +D+ KTAF
Sbjct: 163 RMCIDYRKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAF 342
Query: 1219 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAI-VYIDDVLIFSQSIDQHFKHL 1277
T PF + + MPFGL NA + FQR M IF+ + I V++DD F S +L
Sbjct: 343 TCPFSVFAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANL 522
Query: 1278 NTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1337
+ +K+ + ++ K + LGH I + I + ++ DK P + K
Sbjct: 523 EKVLQRCEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVK-G 699
Query: 1338 LQRFLGCLNYVADFCPQLSTI 1358
+ FLG + + F + +
Sbjct: 700 IHSFLGHVGFYRRFIKDFTKV 762
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 80.1 bits (196), Expect = 7e-15
Identities = 58/201 (28%), Positives = 93/201 (45%), Gaps = 1/201 (0%)
Frame = +1
Query: 1159 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1218
+L I+Y+ LN+A +P+P +L RL + D G+ QI + KD+ K AF
Sbjct: 163 KLCIDYRKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAF 342
Query: 1219 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAI-VYIDDVLIFSQSIDQHFKHL 1277
T PFG + + +PFGL NAP+ FQ M IF + +I V++DD +F S++ K L
Sbjct: 343 TCPFGVFAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKL 522
Query: 1278 NTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1337
+ + + ++ K + LGH I I I+ +K P K
Sbjct: 523 EMVLQRCVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVK-G 699
Query: 1338 LQRFLGCLNYVADFCPQLSTI 1358
++ FLG + F + +
Sbjct: 700 IRSFLGQARFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 75.9 bits (185), Expect = 1e-13
Identities = 62/219 (28%), Positives = 107/219 (48%), Gaps = 3/219 (1%)
Frame = +3
Query: 1154 ERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDR 1213
E G + ++Y+ LN A ++P+P+ L+ FS D SG+ QI++ +D
Sbjct: 12 EDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDM 191
Query: 1214 YKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIF-NP*SKFAIVYIDDVLIFSQSIDQ 1272
KT F +G + + M FGLKN + +QR M +F + K VY+DD+++ S++ ++
Sbjct: 192 EKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEE 371
Query: 1273 HFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIH--QGTIIPINRAIEFTDKFPD 1330
H +L ++K + ++ K +F+ K R L I +G + N+ +
Sbjct: 372 HLVNLRKLFRRLRKYRLRLNPAK-CMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMAKP 548
Query: 1331 QIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKD 1369
+ Q+Q FLG LNY+ F L +PL L K+
Sbjct: 549 H--TEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKN 659
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 67.0 bits (162), Expect = 6e-11
Identities = 39/138 (28%), Positives = 73/138 (52%), Gaps = 1/138 (0%)
Frame = +3
Query: 1159 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1218
R+ ++Y+ LN+A +P+P+ L+A + +FS D SG+ QI++ +D KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 1219 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAI-VYIDDVLIFSQSIDQHFKHL 1277
+G + + VM FGLKN + + R M +F I Y+D+++ S+ ++H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 1278 NTFISIIKKNGMAVSKTK 1295
++K + ++ K
Sbjct: 363 QNLFGQLRKYRLRLNPRK 416
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 59.3 bits (142), Expect = 1e-08
Identities = 44/162 (27%), Positives = 78/162 (47%), Gaps = 1/162 (0%)
Frame = +1
Query: 1338 LQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYL 1396
++ F G ++ F P ST+ PL++ +KK+ W + +K ++ P L L
Sbjct: 121 IRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAPVLAL 300
Query: 1397 PNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLS 1456
P+ +E DAS +G +L Q IA+ S+ + A NY T KE+ A++ +
Sbjct: 301 PDFSKTFELECDASGVGVRAVLLQ----GGHPIAYFSEKLHSATLNYPTYDKELYALIRA 468
Query: 1457 ISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARW 1498
++ L+ ++F++ D +S K I K L +H A+W
Sbjct: 469 PQTWEHFLVCKEFVIHSDHQSLKYIRGK--SKLNKRH--AKW 582
>BE802896
Length = 416
Score = 58.2 bits (139), Expect = 3e-08
Identities = 39/135 (28%), Positives = 68/135 (49%), Gaps = 1/135 (0%)
Frame = -2
Query: 1341 FLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNP 1399
FLG + F + PL + L+K+ ++D +K + P + P+
Sbjct: 412 FLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPDW 233
Query: 1400 QAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISN 1459
A + DAS+ G +L QKI ++I ++S+ + AQ NY+T +KE+LAIV ++
Sbjct: 232 TAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALEK 53
Query: 1460 FQSDLINQKFLVRVD 1474
F S L+ + +V +D
Sbjct: 52 FHSYLLGTRIIVYID 8
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 57.8 bits (138), Expect = 4e-08
Identities = 37/120 (30%), Positives = 62/120 (50%), Gaps = 1/120 (0%)
Frame = +2
Query: 1194 FSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*S 1253
FS D SG+ QI + +D KT F +G + + VM FGLKN + +QR M +F+
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 1254 KFAI-VYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQ 1312
I VY+DD++ S++ +H +L ++K + ++ TK + + LG + Q
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
>TC211261
Length = 520
Score = 47.4 bits (111), Expect = 5e-05
Identities = 25/55 (45%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = -2
Query: 369 RKETKLYYPRATAPDLLLEEKD-SSNFKSFSANNVYEWNIDGENEYGITKILQNM 422
R TKLYY + T PDL L+E+D +S + F + NIDG+ EY T LQ+M
Sbjct: 504 RNSTKLYYQKVTIPDLWLQERDLNSKLQRFRTMGIC**NIDGQIEYNTTHTLQHM 340
>BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, partial (3%)
Length = 153
Score = 42.7 bits (99), Expect = 0.001
Identities = 19/51 (37%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Frame = -1
Query: 1234 LKNAPSEFQRIMNEIF-NP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISI 1283
L NAP+ FQ +MN IF + K+ +V+ DD+L++S + +H HL +
Sbjct: 153 LTNAPTSFQCLMNHIFQHALRKYVLVFFDDILVYSSTWHEHLCHLEVVFKV 1
>BQ627806
Length = 435
Score = 41.6 bits (96), Expect = 0.003
Identities = 23/55 (41%), Positives = 30/55 (53%)
Frame = +3
Query: 1366 LKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQ 1420
L KD W++ Q+KL + P L LP+ + +VETDAS IG G IL Q
Sbjct: 6 LVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQ 170
>AW570005
Length = 413
Score = 40.8 bits (94), Expect = 0.005
Identities = 37/128 (28%), Positives = 53/128 (40%)
Frame = -2
Query: 1338 LQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLP 1397
L+ FL + F + + PL L KD WS + +K + N L LP
Sbjct: 388 LRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVTNTLVLALP 209
Query: 1398 NPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSI 1457
+ VETDAS G +L Q + IAF SK + P ST E+ AI +
Sbjct: 208 DFTKPFTVETDASGSDMGAVLSQ----EGHPIAFFSKEFCPKLVRSSTYVHELAAITNVV 41
Query: 1458 SNFQSDLI 1465
++ L+
Sbjct: 40 KKWRQYLL 17
>TC208012 similar to UP|Q761Z7 (Q761Z7) BRI1-KD interacting protein 117
(Fragment), partial (38%)
Length = 993
Score = 40.0 bits (92), Expect = 0.008
Identities = 47/197 (23%), Positives = 77/197 (38%), Gaps = 12/197 (6%)
Frame = +1
Query: 665 KPRSKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRISELHLEP--EIEDK 722
KP S P A P + P+ CY C K GH+SR C+ ++ LH E E+
Sbjct: 79 KPGSGPSD---APKVPSDAPK----ICYKCKKAGHLSRDCK-EQPDGLLHRNAIGEAEEN 234
Query: 723 INNLLIQTSDEEESVPSDSEVSEDLNQIQNDDDQSSSSINVLTNE---QDLIFRAIDSIP 779
+ I TS + + +D+N+I ++ + + ++ LT D++ A+
Sbjct: 235 PKSTAIDTSQADRVAMEE----DDINEIGEEEKEKLNDVDYLTGNPLPNDILLYAVPVCG 402
Query: 780 DPDEKKVYLERLKLTLEDRPPKSPITTNKFNLKDTF-------KRLEKSTIKPVTIQDLQ 832
+ Y R+K+ + K NL K L K+ P + +
Sbjct: 403 PYSAVQSYKYRVKI-IPGPAKKGKAAKTAMNLFSHMSEATTREKELMKACTDPELVAAIV 579
Query: 833 SEVHILKAEVKSLKQIQ 849
V I A + LKQ Q
Sbjct: 580 GNVKISAAGLTQLKQKQ 630
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 39.3 bits (90), Expect = 0.013
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +2
Query: 671 HSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLK 707
H+++ R G D+ CY CG+PGH +R CR++
Sbjct: 278 HNSRGGGGGRGGRSGGSDLKCYECGEPGHFARECRMR 388
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 39.3 bits (90), Expect = 0.013
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +1
Query: 671 HSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLK 707
H+++ R G D+ CY CG+PGH +R CR++
Sbjct: 187 HNSRGGGGGRGGRSGGSDLKCYECGEPGHFARECRMR 297
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,357,105
Number of Sequences: 63676
Number of extensions: 967443
Number of successful extensions: 9542
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 7608
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9044
length of query: 1526
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1416
effective length of database: 5,635,272
effective search space: 7979545152
effective search space used: 7979545152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0173b.7


