

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173a.1
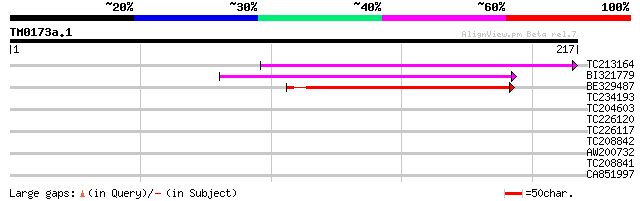
(217 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC213164 81 3e-16
BI321779 77 6e-15
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 72 3e-13
TC234193 39 0.001
TC204603 similar to GB|AAF00023.1|6002520|AF076686 GIGANTEA {Ara... 32 0.22
TC226120 similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24, ... 31 0.49
TC226117 similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24, ... 31 0.49
TC208842 weakly similar to UP|Q9LL85 (Q9LL85) DNA-binding protei... 27 7.1
AW200732 27 7.1
TC208841 weakly similar to UP|Q9LL85 (Q9LL85) DNA-binding protei... 27 7.1
CA851997 27 7.1
>TC213164
Length = 446
Score = 81.3 bits (199), Expect = 3e-16
Identities = 44/121 (36%), Positives = 69/121 (56%)
Frame = +3
Query: 97 MLEVAFTSEEIWEVVIACDGNRALGPYGFNFIFFREFWDIIKDDMMKLFVEFYEFGKLVE 156
+L F +E+ + ++ GP G NF F +EFWDI+K D+++ EFY G ++
Sbjct: 15 LLVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLK 194
Query: 157 GLNLKFIALIPKKKILEAVSVYRPISLTGSIYILIAKDLT*RFHMVMPYLLSWNQFSFTK 216
N F+ALIPK +++ Y+PISL G IY ++AK L+ R V+P ++ Q F +
Sbjct: 195 RSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFME 374
Query: 217 G 217
G
Sbjct: 375 G 377
>BI321779
Length = 421
Score = 77.0 bits (188), Expect = 6e-15
Identities = 41/114 (35%), Positives = 66/114 (56%)
Frame = +2
Query: 81 QLTGDGF*KVKVEDKSMLEVAFTSEEIWEVVIACDGNRALGPYGFNFIFFREFWDIIKDD 140
+L G F ED +ML F EEI +V+ C+ +++ GP G++ + + FW++IK++
Sbjct: 32 KLNGVEFLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEE 211
Query: 141 MMKLFVEFYEFGKLVEGLNLKFIALIPKKKILEAVSVYRPISLTGSIYILIAKD 194
++ F EF++F L G N FIALI K + Y PISL G +Y ++ K+
Sbjct: 212 VINFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*KN 373
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 71.6 bits (174), Expect = 3e-13
Identities = 35/87 (40%), Positives = 54/87 (61%)
Frame = -2
Query: 107 IWEVVIACDGNRALGPYGFNFIFFREFWDIIKDDMMKLFVEFYEFGKLVEGLNLKFIALI 166
+W+ C ++ GP G NF F + FW+ +K D+++ EF+ G +G N FIALI
Sbjct: 363 VWQ----CGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 167 PKKKILEAVSVYRPISLTGSIYILIAK 193
PK K +A++ +RPISL G +Y ++AK
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAK 115
>TC234193
Length = 697
Score = 39.3 bits (90), Expect = 0.001
Identities = 19/64 (29%), Positives = 36/64 (55%)
Frame = -3
Query: 124 GFNFIFFREFWDIIKDDMMKLFVEFYEFGKLVEGLNLKFIALIPKKKILEAVSVYRPISL 183
G+NF F ++ +++K+D+++ EF+ G L G N F+ P + +RP++L
Sbjct: 191 GYNFNFIKKC*EVMKEDVVRAIQEFHSHGCLSRGTNASFLTHPP-----*GLGDFRPVTL 27
Query: 184 TGSI 187
GS+
Sbjct: 26 VGSL 15
>TC204603 similar to GB|AAF00023.1|6002520|AF076686 GIGANTEA {Arabidopsis
thaliana;} , partial (35%)
Length = 1718
Score = 32.0 bits (71), Expect = 0.22
Identities = 14/30 (46%), Positives = 18/30 (59%), Gaps = 2/30 (6%)
Frame = -3
Query: 115 DGNRALGPYGFNFIFFREFW--DIIKDDMM 142
+ N A GP FNF+FF FW D+ K M+
Sbjct: 573 ESNAATGPCFFNFLFFDSFWFSDVCKSSMI 484
>TC226120 similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24, partial
(50%)
Length = 642
Score = 30.8 bits (68), Expect = 0.49
Identities = 15/37 (40%), Positives = 23/37 (61%), Gaps = 4/37 (10%)
Frame = +3
Query: 181 ISLTGSIYILIAKD----LT*RFHMVMPYLLSWNQFS 213
+++ SIYI + K LT F+ +MPYLL W+ F+
Sbjct: 315 VNVDESIYIPVTKAELAVLTSTFNFIMPYLLGWHTFA 425
>TC226117 similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24, partial
(62%)
Length = 1015
Score = 30.8 bits (68), Expect = 0.49
Identities = 15/37 (40%), Positives = 23/37 (61%), Gaps = 4/37 (10%)
Frame = +1
Query: 181 ISLTGSIYILIAKD----LT*RFHMVMPYLLSWNQFS 213
+++ SIYI + K LT F+ +MPYLL W+ F+
Sbjct: 661 VNVDESIYIPVTKAELAVLTSTFNFIMPYLLGWHTFA 771
>TC208842 weakly similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24,
partial (40%)
Length = 984
Score = 26.9 bits (58), Expect = 7.1
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +3
Query: 199 FHMVMPYLLSWNQF 212
F+ +MPYLL WN F
Sbjct: 669 FNCIMPYLLGWNAF 710
>AW200732
Length = 394
Score = 26.9 bits (58), Expect = 7.1
Identities = 19/72 (26%), Positives = 36/72 (49%), Gaps = 5/72 (6%)
Frame = +3
Query: 148 FYEFGKLVEGLNLKFIALIPK---KKILEAVSVYRPISLTGSIYI--LIAKDLT*RFHMV 202
FY F V G +L + L+ + K + + +Y SL + + L++ ++ +F++
Sbjct: 15 FYMFANDVYGFHLSCMVLLNRLSQTKHMSLIKIYTTFSLGYANFWSWLVSVNINNQFYLN 194
Query: 203 MPYLLSWNQFSF 214
+PY S FSF
Sbjct: 195 IPYFASGLLFSF 230
>TC208841 weakly similar to UP|Q9LL85 (Q9LL85) DNA-binding protein p24,
partial (25%)
Length = 653
Score = 26.9 bits (58), Expect = 7.1
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +2
Query: 199 FHMVMPYLLSWNQF 212
F+ +MPYLL WN F
Sbjct: 326 FNYIMPYLLGWNAF 367
>CA851997
Length = 636
Score = 26.9 bits (58), Expect = 7.1
Identities = 13/45 (28%), Positives = 21/45 (45%)
Frame = +1
Query: 122 PYGFNFIFFREFWDIIKDDMMKLFVEFYEFGKLVEGLNLKFIALI 166
P GFN F+ FW + +D+ + + G +N IA+I
Sbjct: 316 PDGFNLGFYHRFWGMCGEDVFQACCMWLAEGAFPSSVNDTTIAII 450
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.346 0.159 0.520
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,598,759
Number of Sequences: 63676
Number of extensions: 100204
Number of successful extensions: 820
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 820
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 820
length of query: 217
length of database: 12,639,632
effective HSP length: 93
effective length of query: 124
effective length of database: 6,717,764
effective search space: 833002736
effective search space used: 833002736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0173a.1


