

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0172.5
(1213 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
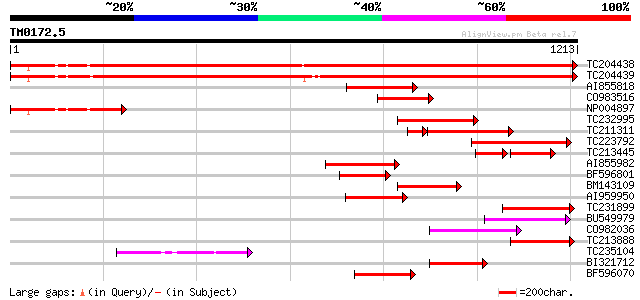
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 1711 0.0
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 1704 0.0
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 216 4e-56
CO983516 214 2e-55
NP004897 gag-protease polyprotein 210 3e-54
TC232995 195 1e-49
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 162 3e-49
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 178 1e-44
TC213445 119 1e-42
AI855982 162 6e-40
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 155 1e-37
BM143109 149 6e-36
AI959950 148 2e-35
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 142 9e-34
BU549979 139 8e-33
CO982036 138 2e-32
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 136 5e-32
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 136 5e-32
BI321712 130 3e-30
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 129 8e-30
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 1711 bits (4431), Expect = 0.0
Identities = 837/1224 (68%), Positives = 1000/1224 (81%), Gaps = 11/1224 (0%)
Frame = +1
Query: 1 ISDEELAEAYKLLYTKWKEACIYGQQQKKQVVELQAEKK-------KLTEEVALLNSKME 53
I+ +ELA +Y+ L K ++ Q KK + L+AEK+ +L EV LNSK+E
Sbjct: 1096 ITFDELAISYRELCIKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLE 1275
Query: 54 GMTKSIRMMSKSTDILDEILDVGKSAGDKTGIGFDYRAVNKASQNMTKEHVQIGRQSNVK 113
MTKSI+M++K +D+LDE+L +GK+ G++ G+GF++++ + + E V +
Sbjct: 1276 NMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSAGRTTMT---EFVPAKNSTGAT 1446
Query: 114 MWNRRPQQSVTHSYAQVRNYQNSTWRCHYCGRFGHIRPYCFKLYGYPILQDVSREYEKKS 173
M R + H Q + + WRCHYCG++GHI+P+C+ L+G+P S S
Sbjct: 1447 MSQHRSR----HHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQS----SSS 1602
Query: 174 PGKSRWKPKVNATALIAHTSLRASSKEDWYFDSGCSRHMTGVKSYLEKMKPHAKSFVTFG 233
K W PK +L+ HTSLRAS+KEDWY DSGCSRHMTGVK +L ++P + S+VTFG
Sbjct: 1603 GRKMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFG 1782
Query: 234 DGAKGKIRGIGKLSDTGSPNLDDVLLVEGLTANLISISQLCDQGLKVVFKQSGCVVKNKN 293
DG+KGKI G+GKL G P+L+ VLLV+GLTANLISISQLCD+G V F +S C+V N+
Sbjct: 1783 DGSKGKITGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEK 1962
Query: 294 KDVLMRGARSKDNCYMWTSETTFLSARCLMSKEDEVRIWHQKLGHLNLKSMKRIVAEEAV 353
+VLM+G+RSKDNCY+WT + T S+ CL SKEDEV+IWHQ+ GHL+L+ MK+I+ + AV
Sbjct: 1963 SEVLMKGSRSKDNCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAV 2142
Query: 354 RGIPKLKIQEGKVCGECQIGKQVKMSHQKLQHLTTSKVLELLHMDLMGPMQVESLGGKRY 413
RGIP LKI+EG++CGECQIGKQVKMSHQKLQH TTS+VLELLHMDLMGPMQVESLGGKRY
Sbjct: 2143 RGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRY 2322
Query: 414 VFVCVDDFSRFTWVEFLKEKSDTFEVFKELCLQVQREKGSGIVKIRSDHGKEFENGQFSE 473
+V VDDFSRFTWV F++EKSDTFEVFKEL L++QREK I +IRSDHG+EFEN +F+E
Sbjct: 2323 AYVVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTE 2502
Query: 474 FCSSEGIKHEFSAPITPQQNGVVERKNRTLQESARAMLHAKNLPYHFWAEAINTACYIHN 533
FC+SEGI HEFSA ITPQQNG+VERKNRTLQE+AR MLHAK LPY+ WAEA+NTACYIHN
Sbjct: 2503 FCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHN 2682
Query: 534 RVTIRQGDTVTQYELWKGKKPTVKYFHVFGSKCYILSDREHRRKLDPKSEVGIFLGYSGN 593
RVT+R+G T YE+WKG+KPTVK+FH+FGS CYIL+DRE RRK+DPKS+ GIFLGYS N
Sbjct: 2683 RVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTN 2862
Query: 594 SRAYRVYNIRTKVMMESINVVVDDTSNERTGQAHDEDDLPYECTNVEPDEPAIQFPNEQE 653
SRAYRV+N RT+ +MESINVVVDD + R E+D+ NV + + +
Sbjct: 2863 SRAYRVFNSRTRTVMESINVVVDDLTPARKKDV--EEDVRTSGDNVADTAKSAENAENSD 3036
Query: 654 NTVSQPPL--ATKEPSIRVQKIHPKENIIGDLNDGVITRSRDL--VSNACFISKVEPKNV 709
+ +P + K PSIR+QK+HPKE IIGD N GV TRSR++ VSN+CF+SK+EPKNV
Sbjct: 3037 SATDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNV 3216
Query: 710 KEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSR 769
KEALTDEFWI +MQEEL QFKRNEVWELVPRP+ NV+GTKWIF+NK++E G +TRNK+R
Sbjct: 3217 KEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKAR 3396
Query: 770 LVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVY 829
LVAQGY+QIEG+DF ETFAPVARLESIRLLLGVAC+LKF+LYQMDVKSAFLNGYLNEE Y
Sbjct: 3397 LVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAY 3576
Query: 830 VEQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKN 889
VEQPKGFVDP+ PDHVYRLKKALYGLKQAPRAWYERLTEFL GY KGGIDKTLFVK++
Sbjct: 3577 VEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQD 3756
Query: 890 GGELMVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFI 949
LM+AQIYVDDIVFGGMSN+M+ FV+QM+SEFEMSLVGELTYFLGLQVKQMED++F+
Sbjct: 3757 AENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFL 3936
Query: 950 TQSKYAKGIVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRP 1009
+QSKYAK IVKKFG+ENA HKRTPA TH+KL+KDE GT VD SLYRSMIGSLLYLTASRP
Sbjct: 3937 SQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRP 4116
Query: 1010 DITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGYCDADWAGS 1069
DIT+AVGVCARYQA PK SHL QVKRI+KY++GTSDYGI+Y H ++S L GYCDADWAGS
Sbjct: 4117 DITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGS 4296
Query: 1070 ADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNV 1129
ADDRKSTSGGCF+L NLISWFSKKQNCVSLSTAEAEYIAAGSSC+QL+WMKQMLKEYNV
Sbjct: 4297 ADDRKSTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNV 4476
Query: 1130 QQDVMTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTEKQLADI 1189
+QDVMTL+CDN+SAINISKNP+QHSRTKHIDIRHH+IR+LV+D +TLEHV TE+Q+ADI
Sbjct: 4477 EQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADI 4656
Query: 1190 FTKALDATQFEKLRQLLGICLFEE 1213
FTKALDA QFEKLR LGICL E+
Sbjct: 4657 FTKALDANQFEKLRGKLGICLLED 4728
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 1704 bits (4413), Expect = 0.0
Identities = 839/1230 (68%), Positives = 1000/1230 (81%), Gaps = 17/1230 (1%)
Frame = +1
Query: 1 ISDEELAEAYKLLYTKWKEACIYGQQQKKQVVELQAEKK-------KLTEEVALLNSKME 53
I+ +ELA +Y+ L K ++ Q KK + +L+AEK+ +L EV LNSK+E
Sbjct: 1093 ITFDELAASYRKLCIKSEKILQQEAQLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLE 1272
Query: 54 GMTKSIRMMSKSTDILDEILDVGKSAGDKTGIGFDYRAVNKASQNMTKEHVQIGRQSNVK 113
MTKSI+M++K +D LDE+L +GK+AG++ G+GF+ ++ + + E V ++
Sbjct: 1273 NMTKSIKMLNKGSDTLDEVLLLGKNAGNQRGLGFNPKSAGRTTMT---EFVPAKNRTGAT 1443
Query: 114 MWNRRPQQSVTHSYAQVRNYQNSTWRCHYCGRFGHIRPYCFKLYGYPILQDVSREYEKKS 173
M R + H Q + + WRCHYCG++GHI+P+C+ L+G+P S KK
Sbjct: 1444 MSQHRSR----HHGMQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSNSRKKM 1611
Query: 174 PGKSRWKPKVNATALIAHTSLRASSKEDWYFDSGCSRHMTGVKSYLEKMKPHAKSFVTFG 233
W PK A +L+ HTSLRAS+KEDWY DSGCSRHMTGVK +L ++P + S+VTFG
Sbjct: 1612 ----MWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFG 1779
Query: 234 DGAKGKIRGIGKLSDTGSPNLDDVLLVEGLTANLISISQLCDQGLKVVFKQSGCVVKNKN 293
DG+KGKI G+GKL G P+L+ VLLV+GLTANLISISQLCD+G V F +S C+V N+
Sbjct: 1780 DGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEK 1959
Query: 294 KDVLMRGARSKDNCYMWTSETTFLSARCLMSKEDEVRIWHQKLGHLNLKSMKRIVAEEAV 353
+VLM+G+RSKDNCY+WT + T S+ CL SKEDEVRIWHQ+ GHL+L+ MK+I+ + AV
Sbjct: 1960 SEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAV 2139
Query: 354 RGIPKLKIQEGKVCGECQIGKQVKMSHQKLQHLTTSKVLELLHMDLMGPMQVESLGGKRY 413
RGIP LKI+EG++CGECQIGKQVKMSHQKLQH TTS+VLELLHMDLMGPMQVESLGGKRY
Sbjct: 2140 RGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRY 2319
Query: 414 VFVCVDDFSRFTWVEFLKEKSDTFEVFKELCLQVQREKGSGIVKIRSDHGKEFENGQFSE 473
+V VDDFSRFTWV F++EKS+TFEVFKEL L++QREK I +IRSDHG+EFEN +F+E
Sbjct: 2320 AYVVVDDFSRFTWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTE 2499
Query: 474 FCSSEGIKHEFSAPITPQQNGVVERKNRTLQESARAMLHAKNLPYHFWAEAINTACYIHN 533
FC+SEGI HEFSA ITPQQNG+VERKNRTLQE+AR MLHAK LPY+ WAEA+NTACYIHN
Sbjct: 2500 FCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHN 2679
Query: 534 RVTIRQGDTVTQYELWKGKKPTVKYFHVFGSKCYILSDREHRRKLDPKSEVGIFLGYSGN 593
RVT+R+G T YE+WKG+KP+VK+FH+FGS CYIL+DRE RRK+DPKS+ GIFLGYS N
Sbjct: 2680 RVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTN 2859
Query: 594 SRAYRVYNIRTKVMMESINVVVDDTSNERTGQAHDE--------DDLPYECTNVEPDEPA 645
SRAYRV+N RT+ +MESINVVVDD S R ++ D N E + A
Sbjct: 2860 SRAYRVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSA 3039
Query: 646 IQFPNEQENTVSQPPLATKEPSIRVQKIHPKENIIGDLNDGVITRSRD--LVSNACFISK 703
E+ ++QP K S R+QK+HPKE IIGD N GV TRSR+ +VSN+CF+SK
Sbjct: 3040 TD-----ESNINQPD---KRSSTRIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSK 3195
Query: 704 VEPKNVKEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNV 763
+EPKNVKEALTDEFWI +MQEEL QFKRNEVWELVPRP+ NV+GTKWIF+NK++E G +
Sbjct: 3196 IEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVI 3375
Query: 764 TRNKSRLVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGY 823
TRNK+RLVAQGY+QIEG+DF ETFAPVARLESIRLLLGVAC+LKF+LYQMDVKSAFLNGY
Sbjct: 3376 TRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGY 3555
Query: 824 LNEEVYVEQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKT 883
LNEEVYVEQPKGF DP+ PDHVYRLKKALYGLKQAPRAWYERLTEFL GY KGGIDKT
Sbjct: 3556 LNEEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKT 3735
Query: 884 LFVKKNGGELMVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQM 943
LFVK++ LM+AQIYVDDIVFGGMSN+M+ FV+QM+SEFEMSLVGELTYFLGLQVKQM
Sbjct: 3736 LFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQM 3915
Query: 944 EDTLFITQSKYAKGIVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLY 1003
ED++F++QS+YAK IVKKFG+ENA HKRTPA TH+KL+KDE GT VD SLYRSMIGSLLY
Sbjct: 3916 EDSIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLY 4095
Query: 1004 LTASRPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGYCD 1063
LTASRPDIT+AVGVCARYQA PK SHL QVKRI+KY++GTSDYGI+Y H +N L GYCD
Sbjct: 4096 LTASRPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCD 4275
Query: 1064 ADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQM 1123
ADWAGSADDRKSTSGGCF+L NNLISWFSKKQNCVSLSTAEAEYIAAGSSC+QL+WMKQM
Sbjct: 4276 ADWAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQM 4455
Query: 1124 LKEYNVQQDVMTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTE 1183
LKEYNV+QDVMTL+CDN+SAINISKNP+QHSRTKHIDIRHH+IR+LV+D +TL+HV TE
Sbjct: 4456 LKEYNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTE 4635
Query: 1184 KQLADIFTKALDATQFEKLRQLLGICLFEE 1213
+Q+ADIFTKALDA QFEKLR LGICL EE
Sbjct: 4636 EQIADIFTKALDANQFEKLRGKLGICLLEE 4725
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 216 bits (551), Expect = 4e-56
Identities = 101/151 (66%), Positives = 125/151 (81%)
Frame = -3
Query: 721 SMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVAQGYSQIEG 780
+MQEEL QF+RN VW+LV +P++ V+GTKW+FRNK DE G + RNK+RLVA+GY+Q EG
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 781 IDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQPKGFVDPS 840
ID+ ET+APVARLE IR+LL ++ F+LYQMDVKSAFLNG + EEVYVEQP GF P
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 841 FPDHVYRLKKALYGLKQAPRAWYERLTEFLI 871
P HVY+L+KALYGLKQAPRAWYER++ FL+
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLL 6
>CO983516
Length = 724
Score = 214 bits (544), Expect = 2e-55
Identities = 103/120 (85%), Positives = 110/120 (90%)
Frame = +2
Query: 787 FAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQPKGFVDPSFPDHVY 846
F PVARLESIRLLLGVAC+LKF+LYQMDVKSAFLNGYLNEEVYVEQPKGF+DP+ PDHVY
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 847 RLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNGGELMVAQIYVDDIVFG 906
RLKKALYGLKQAPRAWYERLTE L GY KGGIDKTLFVK++ LM+AQIYVDDIVFG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>NP004897 gag-protease polyprotein
Length = 1923
Score = 210 bits (534), Expect = 3e-54
Identities = 110/257 (42%), Positives = 159/257 (61%), Gaps = 7/257 (2%)
Frame = +1
Query: 1 ISDEELAEAYKLLYTKWKEACIYGQQQKKQVVELQAEKK-------KLTEEVALLNSKME 53
I+ +ELA +Y+ L K ++ Q KK + L+AEK+ +L EV LNSK+E
Sbjct: 1096 ITFDELATSYRELCIKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLE 1275
Query: 54 GMTKSIRMMSKSTDILDEILDVGKSAGDKTGIGFDYRAVNKASQNMTKEHVQIGRQSNVK 113
MTKSI+M++K +D+LDE+L +GK+ G++ G+GF++++ + + E V +
Sbjct: 1276 NMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSAGRITMT---EFVPAKISTGAT 1446
Query: 114 MWNRRPQQSVTHSYAQVRNYQNSTWRCHYCGRFGHIRPYCFKLYGYPILQDVSREYEKKS 173
M R + H Q + + WRCHYCG++GHI+P+C+ L+G+P S S
Sbjct: 1447 MSQHRSR----HHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQS----SSS 1602
Query: 174 PGKSRWKPKVNATALIAHTSLRASSKEDWYFDSGCSRHMTGVKSYLEKMKPHAKSFVTFG 233
K W PK +L+ HTSLRAS+KEDWY DSGCSRHMTGVK +L ++P + S+VTFG
Sbjct: 1603 RRKMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFG 1782
Query: 234 DGAKGKIRGIGKLSDTG 250
DG+KGKI G+GKL G
Sbjct: 1783 DGSKGKITGMGKLVHDG 1833
>TC232995
Length = 1009
Score = 195 bits (495), Expect = 1e-49
Identities = 89/173 (51%), Positives = 126/173 (72%)
Frame = +2
Query: 830 VEQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKN 889
VEQP GF P+HVY+L+KALYGLKQAPRAWYERL+ FL+ + +G +D TLF+K+
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 890 GGELMVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFI 949
++++ QIYVDDI+FG ++ + ++F M+SEFEMS++GEL YFLGLQ+KQ + +FI
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 950 TQSKYAKGIVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLL 1002
QSKY K ++K+FG+++A H TP +T+ L KDE G +D YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 162 bits (409), Expect(2) = 3e-49
Identities = 83/184 (45%), Positives = 114/184 (61%)
Frame = +3
Query: 894 MVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFITQSK 953
++ IYVDDI+FG S +M ++F E MK FE S+ GEL + LGLQ+ Q +FI Q K
Sbjct: 489 LIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFIHQEK 668
Query: 954 YAKGIVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRPDITF 1013
Y K +K+F ++ A TP + KDEKG Y MI SL YLT+SRPDI F
Sbjct: 669 YTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRPDIVF 848
Query: 1014 AVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGYCDADWAGSADDR 1073
V +CAR+Q+ PK SH+ VKRI++Y+ GT+++ + + + L GYCD +AG +R
Sbjct: 849 VVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFAGDKVER 1028
Query: 1074 KSTS 1077
KSTS
Sbjct: 1029 KSTS 1040
Score = 53.1 bits (126), Expect(2) = 3e-49
Identities = 24/42 (57%), Positives = 30/42 (71%)
Frame = +2
Query: 852 LYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNGGEL 893
+YGLKQA RAWYERL+ FL++NG+ +G D LF K G L
Sbjct: 362 VYGLKQALRAWYERLSSFLVSNGFTRGITDPALFRKAQKGNL 487
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein, partial
(7%)
Length = 804
Score = 178 bits (451), Expect = 1e-44
Identities = 89/218 (40%), Positives = 137/218 (62%), Gaps = 4/218 (1%)
Frame = +1
Query: 989 VDPSLYRSMIGSLLYLTASRPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGI 1048
VD + +R +IGSL YL SRP+I FAV + +R+ P+ SH+ KR+++ I GT G+
Sbjct: 7 VDVTEFRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGV 186
Query: 1049 LYSHNTNSG---LTGYCDADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEA 1105
L+ SG L GY D+DW + KST G F + ++ SKKQ+ ++LST EA
Sbjct: 187 LFPFKAKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEA 366
Query: 1106 EYIAAGSSCTQLMWMKQMLKEYNVQQ-DVMTLFCDNLSAINISKNPIQHSRTKHIDIRHH 1164
EY+AA Q +WM +L+E +++ + L DN SAIN++K+P H R+KHI++R H
Sbjct: 367 EYVAASLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFH 546
Query: 1165 FIRELVEDGTVTLEHVSTEKQLADIFTKALDATQFEKL 1202
+IR+ V G VT+E+ E+QLAD+ TK + ++F+++
Sbjct: 547 YIRDQVSKGNVTVEYCKAEEQLADLMTKPIQVSRFKQI 660
>TC213445
Length = 705
Score = 119 bits (297), Expect(2) = 1e-42
Identities = 57/96 (59%), Positives = 73/96 (75%)
Frame = +1
Query: 1072 DRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNVQQ 1131
DR+STS C F+ + L+SW SKKQN V LSTAEAEYI+A S Q+ WM+Q L +Y ++
Sbjct: 406 DRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYGLKL 585
Query: 1132 DVMTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIR 1167
D + + CDN SAIN+SKN I +SRTKHI+IRHHF+R
Sbjct: 586 DHIPIRCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 74.3 bits (181), Expect(2) = 1e-42
Identities = 35/69 (50%), Positives = 47/69 (67%)
Frame = +2
Query: 996 SMIGSLLYLTASRPDITFAVGVCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTN 1055
+MI S LYL+ SRP I F+V +C RYQA PK SHL +KRI++Y+ G + G+ Y N++
Sbjct: 194 AMIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSS 373
Query: 1056 SGLTGYCDA 1064
L GY DA
Sbjct: 374 YNLVGYSDA 400
>AI855982
Length = 484
Score = 162 bits (411), Expect = 6e-40
Identities = 80/161 (49%), Positives = 113/161 (69%), Gaps = 2/161 (1%)
Frame = +2
Query: 675 PKENIIGDLNDGVITRS--RDLVSNACFISKVEPKNVKEALTDEFWIQSMQEELGQFKRN 732
P +NIIGD++ GV TR +DL +N F+S +EPKN+KEA+ D+ WI +MQEEL QF+RN
Sbjct: 2 PLDNIIGDISKGVTTRHSLKDLCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQFERN 181
Query: 733 EVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVAQGYSQIEGIDFYETFAPVAR 792
VW+LV +PD+ V+ TKW+FRNK DE + +K+RLVA+GY+Q++G+D+ T+A +AR
Sbjct: 182 NVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYASIAR 361
Query: 793 LESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQP 833
L I + L ++ LY SA L+G L EVYV+QP
Sbjct: 362 L*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 155 bits (391), Expect = 1e-37
Identities = 71/111 (63%), Positives = 92/111 (81%)
Frame = +3
Query: 705 EPKNVKEALTDEFWIQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVT 764
EPKN+KEA+ D+ WI MQEEL QF+RN VW+LV +P++ V+GTKW+FRNK DE G +
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 765 RNKSRLVAQGYSQIEGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDV 815
RNK+RLVA+GY+Q EGID+ ET+APVARLE+IR+LL A ++ F+LYQMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>BM143109
Length = 415
Score = 149 bits (377), Expect = 6e-36
Identities = 66/136 (48%), Positives = 103/136 (75%)
Frame = +1
Query: 831 EQPKGFVDPSFPDHVYRLKKALYGLKQAPRAWYERLTEFLINNGYDKGGIDKTLFVKKNG 890
+QP + P+HV++LKK LYGLKQA RAWYE L++FL++ G+ KG +D LF+ K
Sbjct: 1 DQPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKL 180
Query: 891 GELMVAQIYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFIT 950
++++ QIYVDDI+FG ++ + ++F + M++EFEMS++ EL +FLGLQ+KQ ++ +FI+
Sbjct: 181 NDILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFIS 360
Query: 951 QSKYAKGIVKKFGLEN 966
QSKY K ++ +FG+EN
Sbjct: 361 QSKYCKDLIHRFGMEN 408
>AI959950
Length = 466
Score = 148 bits (373), Expect = 2e-35
Identities = 77/132 (58%), Positives = 95/132 (71%)
Frame = -1
Query: 719 IQSMQEELGQFKRNEVWELVPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVAQGYSQI 778
+++MQEEL QF++N V +LV P VVG KWIF NK DE G V R K+RLVA+GYSQ
Sbjct: 397 MKAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQ 218
Query: 779 EGIDFYETFAPVARLESIRLLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQPKGFVD 838
EGID+ +TFA VARLE I +LL A +LYQMDVKSAFLNG + +EVYVEQP GF +
Sbjct: 217 EGIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFEN 38
Query: 839 PSFPDHVYRLKK 850
+ HV++L K
Sbjct: 37 ETLHQHVFKLNK 2
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment), partial
(30%)
Length = 687
Score = 142 bits (358), Expect = 9e-34
Identities = 71/154 (46%), Positives = 100/154 (64%), Gaps = 1/154 (0%)
Frame = +2
Query: 1055 NSGLTGYCDADWAGSADDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSC 1114
N+ L+GYCDADWAG DR+STSG C F+ NL+SW SKKQ V+ S+AEAEY +
Sbjct: 8 NTQLSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVT 187
Query: 1115 TQLMWMKQMLKEYNVQQDV-MTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDG 1173
+LMW+KQ L+E +++ M L+CDN +A++I+ NP+ H RTKHI+I HFIRE +
Sbjct: 188 CELMWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSK 367
Query: 1174 TVTLEHVSTEKQLADIFTKALDATQFEKLRQLLG 1207
+ E + + Q DI TK+L + + + LG
Sbjct: 368 EIVTEFIGSNDQPVDILTKSLRGPKIQIVCSKLG 469
>BU549979
Length = 615
Score = 139 bits (350), Expect = 8e-33
Identities = 66/187 (35%), Positives = 108/187 (57%), Gaps = 3/187 (1%)
Frame = -1
Query: 1017 VCARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNSGLTGYCDADWAGSADDRKST 1076
V RYQ+ P H K++++Y+ GT DY ++Y + GY D+D+AG D R+ST
Sbjct: 615 VLGRYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRST 436
Query: 1077 SGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNVQQDV--- 1133
SG F L + ++SW S KQ ++ ST E E++ + + +W+K + V +
Sbjct: 435 SGYIFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRP 256
Query: 1134 MTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTEKQLADIFTKA 1193
+ L+CDN +A+ ++KN +R+KHIDI++ IRE V++ V +EHV+TE + D TK
Sbjct: 255 LKLYCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDPLTKG 76
Query: 1194 LDATQFE 1200
+ F+
Sbjct: 75 MTPKNFK 55
>CO982036
Length = 674
Score = 138 bits (347), Expect = 2e-32
Identities = 74/201 (36%), Positives = 117/201 (57%), Gaps = 3/201 (1%)
Frame = -2
Query: 898 IYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFITQSKYAKG 957
+YVD I+ G S +++ ++ S F + L+G+L YF+ ++VK M D LF ++ +
Sbjct: 640 VYVD-IIITGSSCTLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDLLFSLRTSIFEI 464
Query: 958 IVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRPDITFAVGV 1017
+K + A +P T KL+K + P+ YRS++G+L Y T RP+I+FAV
Sbjct: 463 FCRKPR*Q-AQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTVIRPEISFAVNK 287
Query: 1018 CARYQAEPKTSHLIQVKRIIKYISGTSDYGILYSHNTNS---GLTGYCDADWAGSADDRK 1074
++ + P SH +VKRI++Y+ G+ YG+ +S + G+CDADWA + DD++
Sbjct: 286 VCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCDADWASAVDDKR 107
Query: 1075 STSGGCFFLENNLISWFSKKQ 1095
STSG FL NLISW+ KQ
Sbjct: 106 STSGAAVFLGPNLISWWXXKQ 44
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 136 bits (343), Expect = 5e-32
Identities = 67/139 (48%), Positives = 95/139 (68%), Gaps = 1/139 (0%)
Frame = +3
Query: 1071 DDRKSTSGGCFFLENNLISWFSKKQNCVSLSTAEAEYIAAGSSCTQLMWMKQMLKEYNV- 1129
DDRKST+G FF+ + +W SKKQ V+LST EAEY+AA S +W++ +LKE +
Sbjct: 9 DDRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELKMP 188
Query: 1130 QQDVMTLFCDNLSAINISKNPIQHSRTKHIDIRHHFIRELVEDGTVTLEHVSTEKQLADI 1189
Q++ M + DN SA+ ++KNP+ H ++KHID R+HFIRE +E V L++V ++ Q ADI
Sbjct: 189 QEEPMEICVDNKSALALAKNPVFHEKSKHIDTRYHFIRECIEKKEVKLKYVMSQDQAADI 368
Query: 1190 FTKALDATQFEKLRQLLGI 1208
FTK L F KLR +LG+
Sbjct: 369 FTKPLKLETFVKLRSMLGV 425
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 136 bits (343), Expect = 5e-32
Identities = 92/292 (31%), Positives = 149/292 (50%), Gaps = 1/292 (0%)
Frame = +2
Query: 229 FVTFGDGAKGKIRGIGKLSDTGSPNLDDVLLVEGLTANLISISQLCD-QGLKVVFKQSGC 287
F+T DG++ GIG +S T S +L+ V+ + G N+ S+SQL + V F +
Sbjct: 17 FITLADGSRVVATGIGHVSPTSSLSLNSVVFILGCPFNITSLSQLTRFRNCSVTFDANSF 196
Query: 288 VVKNKNKDVLMRGARSKDNCYMWTSETTFLSARCLMSKEDEVRIWHQKLGHLNLKSMKRI 347
V++ + Y +++ + K + H++LGH +L +K +
Sbjct: 197 VIQECGTGWTIGVGIESHGLYYLKPNLSWVCSAVTSPK-----LLHERLGHPHLSKLKIM 361
Query: 348 VAEEAVRGIPKLKIQEGKVCGECQIGKQVKMSHQKLQHLTTSKVLELLHMDLMGPMQVES 407
V P L+ + C CQ+GK V+ S + ++ S L ++H D+ GP +V S
Sbjct: 362 V--------PSLEKIKDLFCESCQLGKHVRSSXRHVESRVDSPFL-VIHXDIWGPNRVSS 514
Query: 408 LGGKRYVFVCVDDFSRFTWVEFLKEKSDTFEVFKELCLQVQREKGSGIVKIRSDHGKEFE 467
+ RY +D+FS+ T V +KE+S+ F +++ + G I +RSD+ KE+
Sbjct: 515 MS-YRYFVTFIDEFSQCTRVFLMKERSEILS-FLTSVNKIKTQFGKTIKILRSDNAKEYF 688
Query: 468 NGQFSEFCSSEGIKHEFSAPITPQQNGVVERKNRTLQESARAMLHAKNLPYH 519
+ S F S++GI H+FS P TPQQN + ERKNR L E+AR +L N P H
Sbjct: 689 SSVISPFXSAQGILHQFSCPHTPQQNDIAERKNRHLVETARTLLLHANEPIH 844
>BI321712
Length = 399
Score = 130 bits (328), Expect = 3e-30
Identities = 62/124 (50%), Positives = 90/124 (72%)
Frame = -3
Query: 898 IYVDDIVFGGMSNQMVEQFVEQMKSEFEMSLVGELTYFLGLQVKQMEDTLFITQSKYAKG 957
+YVDD++F G + M E+F + M +EFEM+ +G + Y+LG++VKQ + +FITQ YAK
Sbjct: 379 LYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYAKE 200
Query: 958 IVKKFGLENAGHKRTPAATHIKLTKDEKGTDVDPSLYRSMIGSLLYLTASRPDITFAVGV 1017
++KKF +++A TP KL+K EKG +VDP+LY+S+IGSL YLT +RPDI + VGV
Sbjct: 199 VLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVVGV 20
Query: 1018 CARY 1021
+RY
Sbjct: 19 VSRY 8
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 129 bits (324), Expect = 8e-30
Identities = 60/130 (46%), Positives = 92/130 (70%)
Frame = -2
Query: 738 VPRPDDANVVGTKWIFRNKSDESGNVTRNKSRLVAQGYSQIEGIDFYETFAPVARLESIR 797
VP P VG +W++ K +G V R K+RLVA+GY+Q+ GID+ +TF+PVA+L ++R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 798 LLLGVACLLKFRLYQMDVKSAFLNGYLNEEVYVEQPKGFVDPSFPDHVYRLKKALYGLKQ 857
L L +A + + L+Q+D+K+AFL+G L E++Y+EQP GFV V +L ++LYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 858 APRAWYERLT 867
+PRAW+ + +
Sbjct: 46 SPRAWFGKFS 17
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,583,323
Number of Sequences: 63676
Number of extensions: 709112
Number of successful extensions: 3662
Number of sequences better than 10.0: 149
Number of HSP's better than 10.0 without gapping: 3570
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3610
length of query: 1213
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1105
effective length of database: 5,762,624
effective search space: 6367699520
effective search space used: 6367699520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0172.5


