

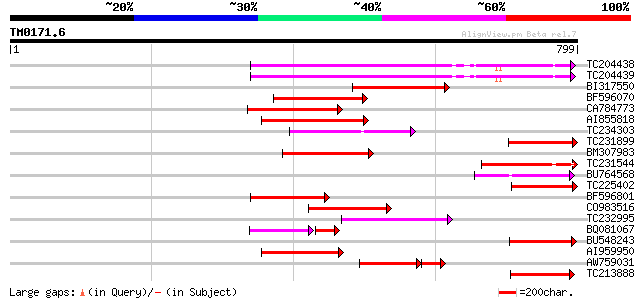
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0171.6
(799 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 262 5e-70
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 261 1e-69
BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Ara... 178 7e-45
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 153 3e-37
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 145 7e-35
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 132 8e-31
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 125 6e-29
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 116 4e-26
BM307983 111 1e-24
TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, ... 103 2e-22
BU764568 102 9e-22
TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotr... 101 1e-21
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 97 3e-20
CO983516 95 1e-19
TC232995 93 4e-19
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 69 4e-19
BU548243 90 3e-18
AI959950 87 3e-17
AW759031 weakly similar to GP|22093573|db polyprotein {Oryza sat... 70 5e-16
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 82 7e-16
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 262 bits (669), Expect = 5e-70
Identities = 163/472 (34%), Positives = 246/472 (51%), Gaps = 14/472 (2%)
Frame = +1
Query: 340 EPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVE 399
EP EA + W++AM E+ + N W LVP P I KW++K K G +
Sbjct: 3202 EPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVIT 3381
Query: 400 RYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDL 459
R KARLVA GY QIEG+ + +TF+P A+L +R++L +A + + L+Q+DV +AFL G L
Sbjct: 3382 RNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYL 3561
Query: 460 YEDVYMKVPEG-VSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSL 518
E+ Y++ P+G V V +L K+ YGLKQA R WY + T L GY++ D +L
Sbjct: 3562 NEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTL 3741
Query: 519 FSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSA 578
F K ++ I IYVDDI+ G + + + F++ +G L YFLGL+V
Sbjct: 3742 FVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQME 3921
Query: 579 KGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTHRKTVL 638
I L Q KY ++V G+ + TP +LS+D G ++ Q +R +
Sbjct: 3922 DSIFLSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSV-----DQSLYRSMIG 4086
Query: 639 SYYNQV*YNLCSSAAKSVPLQSYCDSLCCS-S*NSKVSERESKKRVI--------FPKEF 689
S L +A++ P +Y +C N K+S KR++ + +
Sbjct: 4087 SL-------LYLTASR--PDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMY 4239
Query: 690 C----STIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRAL 745
C S +VG G D R+S + CF++G +LI W SKKQ +S S++EA Y A
Sbjct: 4240 CHCSDSMLVGYCDADWAGSADDRKSTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAA 4419
Query: 746 ASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEI 797
S+ +L + +LK+ +E +YCDN SA++I+ NPV H RTKH++I
Sbjct: 4420 GSSCSQLVWMKQMLKEYNVEQ-DVMTLYCDNMSAINISKNPVQHSRTKHIDI 4572
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 261 bits (666), Expect = 1e-69
Identities = 162/472 (34%), Positives = 246/472 (51%), Gaps = 14/472 (2%)
Frame = +1
Query: 340 EPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVE 399
EP EA + W++AM E+ + N W LVP P I KW++K K G +
Sbjct: 3199 EPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVIT 3378
Query: 400 RYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDL 459
R KARLVA GY QIEG+ + +TF+P A+L +R++L +A + + L+Q+DV +AFL G L
Sbjct: 3379 RNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYL 3558
Query: 460 YEDVYMKVPEG-VSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSL 518
E+VY++ P+G V +L K+ YGLKQA R WY + T L GY++ D +L
Sbjct: 3559 NEEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTL 3738
Query: 519 FSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSA 578
F K ++ I IYVDDI+ G + + + F++ +G L YFLGL+V
Sbjct: 3739 FVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQME 3918
Query: 579 KGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTHRKTVL 638
I L Q +Y ++V G+ + TP +LS+D G ++ Q +R +
Sbjct: 3919 DSIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSV-----DQSLYRSMIG 4083
Query: 639 SYYNQV*YNLCSSAAKSVPLQSYCDSLCCS-S*NSKVSERESKKRVI--------FPKEF 689
S L +A++ P +Y +C N K+S KR++ + +
Sbjct: 4084 SL-------LYLTASR--PDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMY 4236
Query: 690 C----STIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRAL 745
C +VG G D R+S + CF++GN+LI W SKKQ +S S++EA Y A
Sbjct: 4237 CHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAA 4416
Query: 746 ASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEI 797
S+ +L + +LK+ +E +YCDN SA++I+ NPV H RTKH++I
Sbjct: 4417 GSSCSQLVWMKQMLKEYNVEQ-DVMTLYCDNMSAINISKNPVQHSRTKHIDI 4569
>BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Arabidopsis
thaliana}, partial (18%)
Length = 421
Score = 178 bits (452), Expect = 7e-45
Identities = 90/136 (66%), Positives = 108/136 (79%)
Frame = -2
Query: 484 KSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNF 543
KS YGLKQASR+WY K T+LL+ GY Q+ SD+SLF+ T+G +FT LL+YVDDIILAG+
Sbjct: 420 KSLYGLKQASRKWYEKLTNLLLKEGYIQSISDYSLFTLTKGNTFTALLVYVDDIILAGDS 241
Query: 544 LDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKP 603
+DEF RIK LD AFKIK+LG LKYFLGLEV+HS GI++ QRKYCLDL+ DSG+LG KP
Sbjct: 240 IDEFDRIKNVLDLAFKIKNLGKLKYFLGLEVAHSRLGITISQRKYCLDLLKDSGLLGCKP 61
Query: 604 VSTPLDPSSRLSQDGG 619
STPLD S +L G
Sbjct: 60 ASTPLDTSIKLHSAAG 13
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 153 bits (387), Expect = 3e-37
Identities = 75/133 (56%), Positives = 96/133 (71%), Gaps = 1/133 (0%)
Frame = -2
Query: 373 VPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVR 432
VPLPP P+ +WVY +K G V+R KARLVA GY Q+ GI Y DTFSP AKLT VR
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 433 MVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEG-VSCVDSGKVCKLHKSSYGLKQ 491
+ LA+A++ +W LHQLD+ NAFL GDL ED+YM+ P G V+ + G VCKLH+S YGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 492 ASRQWYAKFTSLL 504
+ R W+ KF+ ++
Sbjct: 46 SPRAWFGKFSHVV 8
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 145 bits (366), Expect = 7e-35
Identities = 74/134 (55%), Positives = 89/134 (66%)
Frame = +3
Query: 335 LSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRA 394
LS + P T EA H W AM E+ ALE NGTW LVPLPP + +WVY +K
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 395 NGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAF 454
NG V+R KARLVA GY Q+ GI Y DTFSP LT VR+ LA+A++ +W LHQLD+ NAF
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 455 LLGDLYEDVYMKVP 468
L GDL ED+YM+ P
Sbjct: 363 LHGDLEEDIYMEQP 404
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 132 bits (331), Expect = 8e-31
Identities = 69/151 (45%), Positives = 96/151 (62%), Gaps = 1/151 (0%)
Frame = -3
Query: 356 AMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEG 415
AM E++ E N W LV P N I KWV++ K +G + R KARLVA GYNQ EG
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 416 IYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVSCVD 475
I Y +T++P A+L ++RM+LA S+ N+ L+Q+DV +AFL G + E+VY++ P G D
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 476 S-GKVCKLHKSSYGLKQASRQWYAKFTSLLV 505
V KL K+ YGLKQA R WY + ++ L+
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLL 6
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 125 bits (315), Expect = 6e-29
Identities = 76/180 (42%), Positives = 103/180 (57%), Gaps = 2/180 (1%)
Frame = +1
Query: 395 NGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAF 454
+GT++++KARLVA Y Q+ G Y TFSP AK+ V ++ ++A V +W L LD NAF
Sbjct: 25 SGTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAF 204
Query: 455 LLGDLYEDVYMKVPEG--VSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQA 512
L G L E+VYM+ P G S VC+L +S YGLKQ+ R W F Y
Sbjct: 205 LHGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAW--PFLYCGAAIWYDSH 378
Query: 513 HSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGL 572
+DHS+F Q L++YVDDI + G+ T +K L F+ KDLG L+YFLG+
Sbjct: 379 EADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 116 bits (290), Expect = 4e-26
Identities = 52/97 (53%), Positives = 71/97 (72%)
Frame = +2
Query: 703 GCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDL 762
GC RRS + YC FIG +L+ W+SKKQ +++SS+EA YR++A TCEL + L++L
Sbjct: 47 GCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCELMWIKQFLQEL 226
Query: 763 QIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
+ +YCDNQ+ALHIA+NPVFHERTKH+EI+C
Sbjct: 227 RFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDC 337
>BM307983
Length = 406
Score = 111 bits (277), Expect = 1e-24
Identities = 58/130 (44%), Positives = 82/130 (62%), Gaps = 2/130 (1%)
Frame = +2
Query: 385 KWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAK-LTIVRMVLALASVNNW 443
+W+Y +K A+ T++RYKARLVA GY Q GI Y +TF+ K + + W
Sbjct: 11 RWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQFGW 190
Query: 444 HLHQLDVNNAFLLGDLYEDVYMKVPEGVSCVDSG-KVCKLHKSSYGLKQASRQWYAKFTS 502
+HQ DV NAFL G L E+VYM++P G + G KVC+L K+ YGLKQ+ R W+ +FT
Sbjct: 191 EMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGRFTQ 370
Query: 503 LLVTCGYKQA 512
+++ GYKQ+
Sbjct: 371 AMLSLGYKQS 400
>TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, partial
(16%)
Length = 662
Score = 103 bits (258), Expect = 2e-22
Identities = 62/138 (44%), Positives = 87/138 (62%), Gaps = 3/138 (2%)
Frame = +3
Query: 665 LCCSS*NSKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLIC 724
LC ++ K + +K + F +E I+G C+D+ +S+T YCFF+G+SLI
Sbjct: 12 LCAATRVLKYLKGCPRKGLSFSRESPIQILGFSDADWATCIDSSKSITWYCFFLGSSLIS 191
Query: 725 WRSKKQQTISK--SSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALH- 781
W++KKQ T+S+ SSSEA YRAL S TCEL LTYLLKDL + ++IYCDNQSAL
Sbjct: 192 WKAKKQNTVSRSSSSSEAKYRALTSTTCELQWLTYLLKDLHV-----TLIYCDNQSALQ* 356
Query: 782 IAANPVFHERTKHLEIEC 799
+ ++H + LEI+C
Sbjct: 357 LPIKVIYHGQ---LEIDC 401
>BU764568
Length = 420
Score = 102 bits (253), Expect = 9e-22
Identities = 63/141 (44%), Positives = 83/141 (58%)
Frame = +3
Query: 655 SVPLQSYCDSLCCSS*NSKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSY 714
SVP S SL CS N +++ S KR + + + CRL G T + Y
Sbjct: 3 SVPELSLPGSLECSQLNPEIN*ECSWKRSHLRR*GTFSNYWLFRCRLSGV--T***TSGY 176
Query: 715 CFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYC 774
C IG +LI W+SKKQ ++KSS+EA YRA+A TCEL L LL +L+ E + C
Sbjct: 177 CVLIGGNLISWKSKKQSVVAKSSAEAEYRAMALVTCELIWLKQLL*ELKFEEDTQMTLIC 356
Query: 775 DNQSALHIAANPVFHERTKHL 795
DNQ+ALHIA+NP+FH RTKH+
Sbjct: 357 DNQAALHIASNPIFH*RTKHI 419
>TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotransposon
Hopscotch polyprotein, partial (7%)
Length = 1446
Score = 101 bits (252), Expect = 1e-21
Identities = 46/92 (50%), Positives = 65/92 (70%)
Frame = +2
Query: 708 RRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPV 767
R S YC IG +L+ W+S K +++SS+EA Y+A+ ATCEL + LL++L+
Sbjct: 38 RGSTLGYCVSIGENLVLWKSNK*NVVARSSAEAEYKAMTVATCELIWIKQLLQELKFGST 217
Query: 768 KPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
+ + CDNQ+ALHIA+NPVFHERTKH+EI+C
Sbjct: 218 QQMKLCCDNQAALHIASNPVFHERTKHIEIDC 313
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 97.1 bits (240), Expect = 3e-20
Identities = 50/111 (45%), Positives = 67/111 (60%)
Frame = +3
Query: 340 EPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVE 399
EP EA W+ M E++ E N W LV P N I KWV++ K +G +
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 400 RYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDV 450
R KARLVA GYNQ EGI Y +T++P A+L +RM+LA AS+ N+ L+Q+DV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>CO983516
Length = 724
Score = 94.7 bits (234), Expect = 1e-19
Identities = 49/118 (41%), Positives = 73/118 (61%), Gaps = 1/118 (0%)
Frame = +2
Query: 422 FSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEG-VSCVDSGKVC 480
F P A+L +R++L +A + + L+Q+DV +AFL G L E+VY++ P+G + V
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 481 KLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDII 538
+L K+ YGLKQA R WY + T LL GY++ D +LF K ++ I IYVDDI+
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 718
>TC232995
Length = 1009
Score = 93.2 bits (230), Expect = 4e-19
Identities = 52/157 (33%), Positives = 82/157 (52%), Gaps = 1/157 (0%)
Frame = +2
Query: 468 PEGVSCVDS-GKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQS 526
P G D V KL K+ YGLKQA R WY + ++ L+ + + D +LF K +
Sbjct: 11 PPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRKHND 190
Query: 527 FTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQR 586
++ IYVDDII + + F++ +G LKYFLGL++ + GI + Q
Sbjct: 191 ILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFINQS 370
Query: 587 KYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL 623
KYC +L+ G+ +K +STP+ + L +D G ++
Sbjct: 371 KYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSI 481
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 68.9 bits (167), Expect(2) = 4e-19
Identities = 37/90 (41%), Positives = 50/90 (55%)
Frame = +1
Query: 339 SEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTV 398
+EP T +A W AM + AL N T +L LP ID KWV++IK GT+
Sbjct: 28 AEPSTVKQALISPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFRIKENLYGTL 207
Query: 399 ERYKARLVA*GYNQIEGIYYFDTFSPTAKL 428
RY++RLVA G++ G Y +TFSP +L
Sbjct: 208 NRYRSRLVAKGFHLKFGCDYSETFSPVIEL 297
Score = 44.7 bits (104), Expect(2) = 4e-19
Identities = 19/35 (54%), Positives = 27/35 (76%)
Frame = +3
Query: 431 VRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYM 465
+R++L +A N+W L Q+D+NNAFL G L E+VYM
Sbjct: 303 IRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYM 407
>BU548243
Length = 599
Score = 90.1 bits (222), Expect = 3e-18
Identities = 48/94 (51%), Positives = 63/94 (66%)
Frame = -1
Query: 705 VDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQI 764
VD RS F+G +LI W S+KQQ ++SS+EA YR++A + EL + LL +LQI
Sbjct: 566 VDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIAQTSAELTWIQALLMELQI 387
Query: 765 EPVKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
P P VI CDN+SA+ IA N VFH RTKH+EI+
Sbjct: 386 -PFTPPVILCDNKSAVAIAHNLVFHSRTKHMEID 288
>AI959950
Length = 466
Score = 87.0 bits (214), Expect = 3e-17
Identities = 47/115 (40%), Positives = 71/115 (60%)
Frame = -1
Query: 356 AMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEG 415
AM E+ + N LV LP + KW++ K +G V RYKARLVA GY+Q EG
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 416 IYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEG 470
I Y TF+ A+L ++ ++L+ A+ +N L+Q+DV +AFL G + ++VY++ P G
Sbjct: 211 IDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPG 47
>AW759031 weakly similar to GP|22093573|db polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 430
Score = 69.7 bits (169), Expect(2) = 5e-16
Identities = 35/88 (39%), Positives = 54/88 (60%), Gaps = 1/88 (1%)
Frame = +3
Query: 494 RQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTI-LLIYVDDIILAGNFLDEFTRIKA 552
R W+ +F+S L+ + +DHS+FS I L++Y+DDI++ G+ D R+K+
Sbjct: 6 RAWFGRFSSELLQFCMTRYEADHSVFSLHSPSGLCIYLVVYLDDIVITGDDFDGIHRLKS 185
Query: 553 ALDNAFKIKDLGVLKYFLGLEVSHSAKG 580
L N F+ KDLG YFLG+EV+ S+ G
Sbjct: 186 HLHNKFQTKDLGPPNYFLGIEVAQSSSG 269
Score = 33.5 bits (75), Expect(2) = 5e-16
Identities = 14/34 (41%), Positives = 24/34 (70%)
Frame = +2
Query: 581 ISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRL 614
IS+ QRKY LD++ ++ +L + TP+DP+ +L
Sbjct: 269 ISISQRKYVLDILIETSMLDFRHSDTPMDPNIKL 370
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 82.4 bits (202), Expect = 7e-16
Identities = 40/91 (43%), Positives = 58/91 (62%)
Frame = +3
Query: 706 DTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIE 765
D R+S T + FF+G++ W SKKQ ++ S+ EA Y A S C L LLK+L++
Sbjct: 9 DDRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELKMP 188
Query: 766 PVKPSVIYCDNQSALHIAANPVFHERTKHLE 796
+P I DN+SAL +A NPVFHE++KH++
Sbjct: 189 QEEPMEICVDNKSALALAKNPVFHEKSKHID 281
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.338 0.145 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,986,565
Number of Sequences: 63676
Number of extensions: 608966
Number of successful extensions: 6458
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 6234
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6390
length of query: 799
length of database: 12,639,632
effective HSP length: 105
effective length of query: 694
effective length of database: 5,953,652
effective search space: 4131834488
effective search space used: 4131834488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0171.6


