

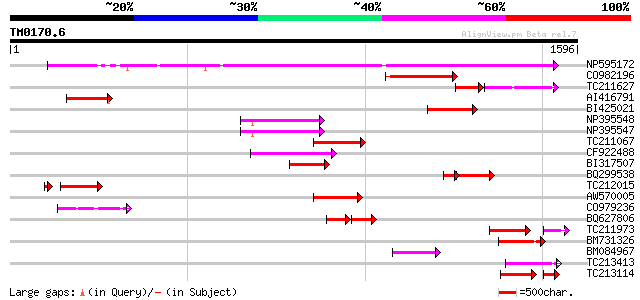
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0170.6
(1596 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 1136 0.0
CO982196 207 4e-53
TC211627 130 5e-51
AI416791 179 7e-45
BI425021 168 2e-41
NP395548 reverse transcriptase [Glycine max] 167 3e-41
NP395547 reverse transcriptase [Glycine max] 160 3e-39
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 156 6e-38
CF922488 149 7e-36
BI317507 144 3e-34
BQ299538 143 7e-34
TC212015 119 1e-31
AW570005 132 1e-30
CO979236 127 3e-29
BQ627806 75 4e-29
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 99 2e-27
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 119 1e-26
BM084967 115 1e-25
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 111 2e-24
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 86 5e-23
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 1136 bits (2938), Expect = 0.0
Identities = 618/1462 (42%), Positives = 869/1462 (59%), Gaps = 22/1462 (1%)
Frame = +1
Query: 106 NEFRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSL 165
+ F+ K++ P FDGK++ WI +AE +F T ++ +A + ++ + +Y L
Sbjct: 271 SSFQVRSVKLDFPRFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQML 450
Query: 166 LATEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPE 225
TE +W+ F AL +G + L +L Q V EY F L ++ L
Sbjct: 451 QKTEPFSSWQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTALVNRVDGLSA 630
Query: 226 KQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGG 285
+ F+ GL+EEI V+++ R L A+ K + ++
Sbjct: 631 EAILDCFVSGLQEEISRDVKAM-------EPRTLTKAVALAKLFE----------EKYTS 759
Query: 286 NGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKN-------TGDIRRAG 338
FS + T T P +N + D K T +
Sbjct: 760 PPKTKTFSNLARNF---------TSNTSATQKYPPTNQKNDNPKPNLPPLLPTPSTKPFN 912
Query: 339 PRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEE-DGEELDESKML 397
R++ +S ++ RR+K LC+ C + H CP++ + LL LEE D ++ DE M+
Sbjct: 913 LRNQNIKKISPAEIQLRREKNLCYFCDEKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMV 1092
Query: 398 AMEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDC 457
E N D++ L+ M + G+ T++ G + G+ V +LVD G++ NF+
Sbjct: 1093 TEEANMDDDTHHLSLNAMR------GSNGVG-TIRFTGQVGGIAVKILVDGGSSDNFIQP 1251
Query: 458 FLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVL 517
+ + L V P + V +G+G A+G L + + ++ L + G D++L
Sbjct: 1252 RVAQVLKLPVEPAPNLRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVIL 1431
Query: 518 GIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQG------MDTRGEHMEALQSITATGERK 571
G WL TLG + ++ + F+ + K+ITLQG + H LQ+ + E
Sbjct: 1432 GSTWLATLGPHVADYAALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECF 1611
Query: 572 P-GLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQE 630
L+ E K LP I+ EL LL+ + VF LPP R ++H I +++
Sbjct: 1612 AIQLIQKEVPEDTLKDLPTNIDP----ELAILLHTYAQVFAVPASLPPQREQDHAIPLKQ 1779
Query: 631 GKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDY 690
G GPV VRPYRYPH K++IEK ++EML GII+ S S +S P++LVKKKD +W C DY
Sbjct: 1780 GSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDY 1959
Query: 691 RALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGH 750
RALN++TV D FP+P ++ELLDELHGA +FSKLDL+SGYHQ+ V+ ED KTAFRTH GH
Sbjct: 1960 RALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGH 2139
Query: 751 YEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEV 810
YE++VMPFGL NAP+TFQ LMN++F+ LR+ VLVFFDDIL+YS +W DH+ HLE VL+
Sbjct: 2140 YEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQT 2319
Query: 811 LQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGL 870
L+ HQL A KC F V+YLGH +S GV+++ KV +V WP P NVK +RGFLGL
Sbjct: 2320 LKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGL 2499
Query: 871 TGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFR 930
TGYYR+FI++Y IA PLT+L +KD+F W +A+ AF LK+ +T APVL+LPDFSQ F
Sbjct: 2500 TGYYRRFIKSYANIAGPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFI 2679
Query: 931 LECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRK 990
LE DASGIG+GA+L HPIAYFSK L PR +SAY +EL+A+ A+ +R YLLG K
Sbjct: 2680 LETDASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNK 2859
Query: 991 FVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGEL 1050
F++ TDQRSLK + Q + T +QQ W K LGY+F+I YKPGK NQ ADALSR+
Sbjct: 2860 FIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFMLA-- 3033
Query: 1051 RPLVSYPLWEEQGVIADENQK-----DPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRV 1105
W E I E + DP L Q++ A Y V +G L ++DRV
Sbjct: 3034 --------WSEPHSIFLEELRARLISDPHLKQLMETYKQGADA-SHYTVREGLLYWKDRV 3186
Query: 1106 VISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRH 1165
VI + ++ K+L+E+H++P GGH+G RT R+ YW M + ++ ++ C +CQ+
Sbjct: 3187 VIPAEEEIVNKILQEYHSSPIGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQA 3366
Query: 1166 KYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHP 1225
K + PAGLLQPLPIP++VWED++++FI GLP S G I VV+DRLTKY+HFIPLK
Sbjct: 3367 KSNNTLPAGLLQPLPIPQQVWEDVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKAD 3546
Query: 1226 YTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDG 1285
Y ++ VAE F I++LHG+P SI+SDRD +F S FW+ +F+LQGT MSSAYHP+SDG
Sbjct: 3547 YNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDG 3726
Query: 1286 QTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLT 1345
Q+E++N+CLE YLRCF + PK W L WAE+W+NT+YH + TPF A+YGR+PP LT
Sbjct: 3727 QSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLT 3906
Query: 1346 RWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKI 1405
R V + L DRD L +LK +L AQ+ M+ QA+ KR F++G+ V VK+
Sbjct: 3907 RQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKL 4086
Query: 1406 RAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGN 1465
+ +RQ S R + KL+ RYFGP+ ++ ++G VAY+L+LP ARIHPVFH+S LK G
Sbjct: 4087 QPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLKPFNGT 4266
Query: 1466 YVEEPGLPEQLDGEEL-PSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLAT 1524
++P LP L E+ P +QP +LA R I+R I Q+LVQW+ +EATWED+
Sbjct: 4267 -AQDPYLPLPLTVTEMGPVMQPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIED 4443
Query: 1525 IRSEFPVFNLEDKVV-SSEGSI 1545
I++ +P FNLEDKVV EG++
Sbjct: 4444 IKASYPTFNLEDKVVFKGEGNV 4509
>CO982196
Length = 812
Score = 207 bits (526), Expect = 4e-53
Identities = 102/203 (50%), Positives = 141/203 (69%)
Frame = +1
Query: 1059 WEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLL 1118
WEE E Q +L +I + PGY + G L ++DR+V+S S+ IP LL
Sbjct: 220 WEE------EIQAYLELYEIYQGILTKTTKKPGYAIRGGKLYFKDRLVLSKNSTKIPLLL 381
Query: 1119 EEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQP 1178
+E +P GGHSGF RT++R+A ++W+GM KT +++V AC++C+R+K SPAGLL
Sbjct: 382 KELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEICRRNKTSTLSPAGLL*L 561
Query: 1179 LPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKE 1238
LPIP +VW DIS++FI GLP+++G + I VVVDRLTKY+HF L HPYTA+ VAE+F KE
Sbjct: 562 LPIPTKVWTDISMDFIGGLPKAQGKDNILVVVDRLTKYAHFFALSHPYTAKEVAELFIKE 741
Query: 1239 IIRLHGVPSSIISDRDPLFVSHF 1261
++RLHG P+SI+SD LF+S F
Sbjct: 742 LVRLHGFPASIVSDXXRLFMSLF 810
Score = 30.8 bits (68), Expect = 5.0
Identities = 13/35 (37%), Positives = 23/35 (65%)
Frame = +2
Query: 983 RPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWA 1017
R Y +G+KF++ T+ RS K Q++++ +Q WA
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKWA 157
>TC211627
Length = 1034
Score = 130 bits (327), Expect(2) = 5e-51
Identities = 77/210 (36%), Positives = 111/210 (52%)
Frame = +3
Query: 1336 VYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEF 1395
+YG+ PP L + G + VEAV+ L L L Q+ M+ A+S R+ F
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1396 EVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFH 1455
+G+WV+V++ +RQ S+ + + KL+ R++GPY I RVG VAYRL+LP ++IHP+FH
Sbjct: 423 NIGDWVYVRL*PYRQTSIQS-TYTKLSKRFYGPYQIQARVGQVAYRLQLPPTSKIHPIFH 599
Query: 1456 ISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPE 1515
+SLLK G E P +QP L + I QVLVQW PE
Sbjct: 600 VSLLKVHHGPIPPELLALPPFSTTNHPLVQPLQFLDWKMDESTTPPIPQVLVQWTNLAPE 779
Query: 1516 EATWEDLATIRSEFPVFNLEDKVVSSEGSI 1545
+ TWE ++ +++LEDKV G I
Sbjct: 780 DTTWESWTQLKD---IYDLEDKVCFQTGGI 860
Score = 91.3 bits (225), Expect(2) = 5e-51
Identities = 39/79 (49%), Positives = 54/79 (67%)
Frame = +2
Query: 1256 LFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHW 1315
+F+S W E+F + GT+ + S+AYHP++DGQTE++NR LE YLR F D P+ W +L
Sbjct: 2 IFISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSL 181
Query: 1316 AEYWFNTSYHSATQQTPFE 1334
AE +NTS HS +PFE
Sbjct: 182 AE*CYNTSVHSGIGFSPFE 238
>AI416791
Length = 420
Score = 179 bits (455), Expect = 7e-45
Identities = 88/138 (63%), Positives = 109/138 (78%), Gaps = 8/138 (5%)
Frame = -3
Query: 159 IHFYNSLLATEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTA 218
IHF+NSL+ +E+LTWE + ALLERYGG+GDGDVYEQL+EL+Q+G VE+YIT+FEYL A
Sbjct: 418 IHFFNSLIGEDEDLTWESLKIALLERYGGHGDGDVYEQLTELKQEGSVEDYITEFEYLIA 239
Query: 219 QIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVA--- 275
QIP+LPEKQ+QGYFLHGL+ EIRG+VR+L MG ++R +LL VTRAVEKEVKG +
Sbjct: 238 QIPRLPEKQFQGYFLHGLQSEIRGRVRTLATMGEMSRTKLLQVTRAVEKEVKGGNGSGFG 59
Query: 276 -----GQARANRFGGNGS 288
G R+N GGN S
Sbjct: 58 RGPKNGPYRSNSHGGNNS 5
>BI425021
Length = 426
Score = 168 bits (425), Expect = 2e-41
Identities = 75/142 (52%), Positives = 102/142 (71%)
Frame = -1
Query: 1176 LQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIF 1235
L PLP+P+R WED+S++FI+GLP G IFVVV+R +K H L +TA VA +F
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLF 247
Query: 1236 AKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLE 1295
+I+LHG P SI+SDRDPLF+SHFW+++FRL GT +MSSAYHP++DGQTE++NR +E
Sbjct: 246 LNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIE 67
Query: 1296 TYLRCFAADQPKTWASWLHWAE 1317
YLR F +P+ ++ W E
Sbjct: 66 QYLRAFVHGRPRNLGRFIPWVE 1
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 167 bits (424), Expect = 3e-41
Identities = 91/254 (35%), Positives = 140/254 (54%), Gaps = 19/254 (7%)
Frame = +1
Query: 650 IEKQVREMLSAGIIRQ-STSAYSSPVILVKKKD------------------QTW*MCVDY 690
+ K+V ++L G+I S SA+ SPV++V KK+ +W +C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 691 RALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGH 750
R LN T D FP+P ++++L+ L G ++ LD GY+Q+ V +D K AF G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 751 YEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEV 810
+ Y +PFGL NAP+TFQ M +F ++ + + VF DD V+ + + LE VL+
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 811 LQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGL 870
L N +KC F + LGH IS +G+ VD K+ ++ P P NVKG+R FLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 871 TGYYRKFIQNYGKI 884
+YR+FI+++ K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 160 bits (406), Expect = 3e-39
Identities = 91/254 (35%), Positives = 139/254 (53%), Gaps = 19/254 (7%)
Frame = +1
Query: 650 IEKQVREMLSAGIIRQ-STSAYSSPVILVKKKD------------------QTW*MCVDY 690
+ K+V ++L AG+I S S++ SPV +V KK W MC+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 691 RALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGH 750
R LN T D +P+P ++++L L F+ LD SGY+Q+ V +D KTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 751 YEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEV 810
+ Y MPFGL NA +TFQ M +F M+ + + VF DD + ++ + + +LE+VL+
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 811 LQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGL 870
+ L N +KC F + LGH IS++G+ V K+ + P P NVKG+ FLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 871 TGYYRKFIQNYGKI 884
G+YR+FI+++ K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 156 bits (395), Expect = 6e-38
Identities = 78/147 (53%), Positives = 103/147 (70%), Gaps = 1/147 (0%)
Frame = +1
Query: 856 PEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKD-AFNWGKQAQQAFEDLKEKL 914
P K+V +R F GL +YR+F+ N+ +A PL EL KK+ AF WG++ +QAF LKEKL
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 915 TTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMA 974
T APVLALPDFS+ F LECDASG+G+ A+LL G HPIAYFS+ L L Y+KEL A
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 975 VALAIQHWRPYLLGRKFVVSTDQRSLK 1001
+ A Q W +L+ ++FV+ +D +SLK
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLK 537
Score = 32.3 bits (72), Expect = 1.7
Identities = 12/42 (28%), Positives = 22/42 (51%)
Frame = +2
Query: 825 FATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRG 866
F + + G ++ GV +DP K+ +++ WP P V + G
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEILG 127
>CF922488
Length = 741
Score = 149 bits (377), Expect = 7e-36
Identities = 91/245 (37%), Positives = 132/245 (53%), Gaps = 1/245 (0%)
Frame = +3
Query: 677 VKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVRE 736
V K+D *MCVDYR LN + DKFP+P I L+D FS +D SGY+Q+++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 737 EDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKT 796
ED+ KT F T G + Y M FGL N +T+Q M +F M+ + + V+ DD++V S+T
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 797 WSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWP 856
+H+++L ++ L+ ++L N KC F K + L + S +G+ VD NKV +
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 857 EPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAF-NWGKQAQQAFEDLKEKLT 915
+P K V+GFLG Y +FI PL L K+ F W AFE +K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 916 TAPVL 920
VL
Sbjct: 723 NPHVL 737
>BI317507
Length = 359
Score = 144 bits (363), Expect = 3e-34
Identities = 69/115 (60%), Positives = 86/115 (74%), Gaps = 1/115 (0%)
Frame = -1
Query: 787 FDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDP 846
F +IL+YS W H++HL VL+VL+ +L AN+KKC F+ +EYLGH+IS+ VA+D
Sbjct: 359 FYNILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDS 180
Query: 847 NKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIA-RPLTELTKKDAFNWG 900
NKV SV WP PKNVK V FL LTGYYRKFI++YGK+A RPLT+LTK D F WG
Sbjct: 179 NKVKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKWG 15
>BQ299538
Length = 426
Score = 143 bits (360), Expect = 7e-34
Identities = 66/109 (60%), Positives = 81/109 (73%)
Frame = +3
Query: 1257 FVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWA 1316
F + + +L GT KMS++YHP DGQT VN CLET+LRCF ADQPK WL WA
Sbjct: 105 FCEFILEGIVKLPGTYLKMSTSYHP*IDGQT--VNHCLETFLRCFVADQPKM*VQWLSWA 278
Query: 1317 EYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRD 1365
EYW+NT++H++T TPFE VYGRKPPVL R++ GE RVEAV ++LQDRD
Sbjct: 279 EYWYNTNFHASTGTTPFEVVYGRKPPVLNRFLPGEVRVEAVRRELQDRD 425
Score = 74.3 bits (181), Expect = 4e-13
Identities = 30/44 (68%), Positives = 39/44 (88%)
Frame = +1
Query: 1222 LKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEM 1265
LKHPY+AR +AEIF KE++ LHGVP+S++SD DP+FVS FWKE+
Sbjct: 1 LKHPYSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>TC212015
Length = 659
Score = 119 bits (297), Expect(2) = 1e-31
Identities = 57/118 (48%), Positives = 84/118 (70%)
Frame = -2
Query: 142 SPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLERYGGNGDGDVYEQLSELR 201
+P+ + L QL ++G TIHF+ SLL LTWE+ + LLE YGG +GD E+L+ +R
Sbjct: 433 APKCEGELGQLFLDGSTIHFFKSLLDEYPSLTWEKLKSELLE*YGGIDEGDDLERLAVIR 254
Query: 202 QQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLL 259
Q G+V++YI +FE LTAQ +LP Q+ GYF+HGLK+ IRG+VRSL +G + ++R++
Sbjct: 253 QDGMVDKYILEFETLTAQESRLPNDQFFGYFVHGLKDGIRGRVRSLHTLGPLFQSRMM 80
Score = 37.7 bits (86), Expect(2) = 1e-31
Identities = 16/22 (72%), Positives = 20/22 (90%)
Frame = -1
Query: 99 VLQGEALNEFRQSVKKVELPMF 120
+L G+ L+EFR+SVKKVELPMF
Sbjct: 560 ILHGDTLDEFRKSVKKVELPMF 495
Score = 35.0 bits (79), Expect = 0.26
Identities = 18/31 (58%), Positives = 20/31 (64%)
Frame = -3
Query: 123 KDLAGWISRAEIYFKVQETSPEVKVSLAQLS 153
+D AGWI E+YF VQ T P VKVS A S
Sbjct: 489 EDPAGWIICVEVYF*VQGTHPNVKVS*ASCS 397
>AW570005
Length = 413
Score = 132 bits (332), Expect = 1e-30
Identities = 64/137 (46%), Positives = 87/137 (62%)
Frame = -2
Query: 856 PEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLT 915
P P+ + +RGFL LTG+YR+FI+ Y +A PL+ L KD+F W +A AF+ LK +T
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVT 233
Query: 916 TAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAV 975
VLALPDF++ F +E DASG +GA+L HPIA+FSK P+ + S Y EL A+
Sbjct: 232 NTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAI 53
Query: 976 ALAIQHWRPYLLGRKFV 992
++ WR YLLG V
Sbjct: 52 TNVVKKWRQYLLGHHLV 2
>CO979236
Length = 729
Score = 127 bits (320), Expect = 3e-29
Identities = 87/210 (41%), Positives = 110/210 (51%), Gaps = 2/210 (0%)
Frame = -1
Query: 136 FKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLERYGGNGDGDVYE 195
F+VQETS EVKV LAQ+SME GT+HF+N LL E LTWE + L++RY G G YE
Sbjct: 729 FQVQETSDEVKVRLAQISMENGTVHFFNLLLLENENLTWEELKYELMQRYAGK--GTAYE 556
Query: 196 QLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNR 255
QLS L+Q +++YI FE L AQIPKL ++QY F HGLK SL + R
Sbjct: 555 QLSALQQTRSIDDYIQRFECLIAQIPKLQDEQYFACFTHGLK--------SLHVANLLTR 400
Query: 256 ARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSR--SGFSGATKGNGTEWIWVKGNKETG 313
RL+ + RAV+ E+ A R + +G+R SG G G K T
Sbjct: 399 GRLMNIARAVKMEMTSKNRAWVGRGDTCMESGTRTASGQRNPLVATGRTGSRNYGGKGT- 223
Query: 314 QTVNRPNSNPRGDQTKNTGDIRRAGPRDRG 343
N P N RGD + + G DRG
Sbjct: 222 ---NGPGPNARGDGAEK----YKQGQ*DRG 154
>BQ627806
Length = 435
Score = 74.7 bits (182), Expect(2) = 4e-29
Identities = 36/68 (52%), Positives = 46/68 (66%)
Frame = +3
Query: 891 LTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHP 950
L KD F+W ++A +AF LK L APVL LPDF+ F +E DASGIG+GAIL HP
Sbjct: 3 LLVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHP 182
Query: 951 IAYFSKAL 958
+A+F A+
Sbjct: 183 LAFFIYAI 206
Score = 73.6 bits (179), Expect(2) = 4e-29
Identities = 33/71 (46%), Positives = 49/71 (68%)
Frame = +1
Query: 963 LAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLG 1022
L S Y +EL A+ +A++ WR YLLG FV+ TD RSLKE + Q + T +QQ + A+L+G
Sbjct: 220 LRASTYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMG 399
Query: 1023 YNFEIIYKPGK 1033
+++ I Y+ GK
Sbjct: 400 FDYTIQYRAGK 432
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 99.0 bits (245), Expect(2) = 2e-27
Identities = 48/114 (42%), Positives = 78/114 (68%)
Frame = +1
Query: 1352 TRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQV 1411
+R+E V K + RD L L+++L+ + + M A N +R+ E+ VG+ VF+K++ +R+
Sbjct: 7 SRLEEVNKLIIARDGLLATLRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYRRR 186
Query: 1412 SLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGN 1465
SLA R++ KL+ R++ P+ + +VG +AY+L LP +IHPVFH+SLLKKA N
Sbjct: 187 SLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHIKIHPVFHVSLLKKASPN 348
Score = 43.5 bits (101), Expect(2) = 2e-27
Identities = 23/72 (31%), Positives = 38/72 (51%)
Frame = +3
Query: 1504 QVLVQWQGKTPEEATWEDLATIRSEFPVFNLEDKVVSSEGSIVRKDNNHNGLLAQDDPGP 1563
+VL+QW+ P E +WE +A ++ F +++LEDKV G I + + P
Sbjct: 438 KVLIQWKNLPPSENSWESVAKLQEIFSIYHLEDKVSLLGGGIDKHKHK-----------P 584
Query: 1564 RVWKVYSRRGQR 1575
+ KVY+R+ R
Sbjct: 585 PIPKVYTRKHPR 620
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 119 bits (297), Expect = 1e-26
Identities = 60/134 (44%), Positives = 89/134 (65%)
Frame = +1
Query: 1375 LVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGR 1434
L AQ M AN+ R+ + VG+WV++KIR HRQ S+ R+H KL AR++GPY ++ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1435 VGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRD 1494
VGAVA++L+LP ARIHPVFH+S LK+A+GN+ + LP L+ + P +L R+
Sbjct: 205 VGAVAFQLQLPSEARIHPVFHVSQLKRALGNHQAQEELPPDLE-HQAELYFPVQILKIRE 381
Query: 1495 ILRQGESISQVLVQ 1508
+ +Q E QVL++
Sbjct: 382 VQKQHEVERQVLIR 423
>BM084967
Length = 426
Score = 115 bits (289), Expect = 1e-25
Identities = 58/136 (42%), Positives = 81/136 (58%)
Frame = -2
Query: 1077 QIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTY 1136
Q+ + +P +P + V+Q +L + + + S IP LL E+H++P H G +T
Sbjct: 413 QLQQAILAEPTTYPDHSVIQDLILKNGCIWLPSGFSFIPTLLLEYHSSPTDAHIGVTKTM 234
Query: 1137 RRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIG 1196
R++ N W G+ K +++FV AC CQ KY A AGLL PLP+P R WED+S NFIIG
Sbjct: 233 ARLSENFTWIGIRKDVEQFVAACLDCQYTKYEAQKMAGLLCPLPVPCRPWEDLSFNFIIG 54
Query: 1197 LPRSRGFEAIFVVVDR 1212
L RG+ AI VVV R
Sbjct: 53 LSEFRGYTAILVVVGR 6
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 111 bits (278), Expect = 2e-24
Identities = 62/156 (39%), Positives = 88/156 (55%)
Frame = +1
Query: 1397 VGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHI 1456
+G+WV VK+R HRQ S + ++KL RY+GP+ + R+G V YRLKL +RIHPVFH+
Sbjct: 4 IGDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAHSRIHPVFHV 183
Query: 1457 SLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEE 1516
SLLK VG+ P + E + P V+ + + VLVQW + ++
Sbjct: 184 SLLKAFVGDPETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRMVLVQWPSASRQD 363
Query: 1517 ATWEDLATIRSEFPVFNLEDKVVSSEGSIVRKDNNH 1552
A+WED +R +NLEDKV+S E R D+ H
Sbjct: 364 ASWEDWQVLRER---YNLEDKVLSEE----RGDDTH 450
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 86.3 bits (212), Expect(2) = 5e-23
Identities = 45/102 (44%), Positives = 64/102 (62%)
Frame = +3
Query: 1382 MRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYR 1441
M+A A+ R+ VG+WV++K++ +R SLA + + KL+ R++GPY I ++G VA+
Sbjct: 3 MKAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFE 182
Query: 1442 LKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPS 1483
L LP +IHPVFH SLLKKAV LP L E+L S
Sbjct: 183 LDLPPARKIHPVFHASLLKKAVAATANPQPLPLML-SEDLSS 305
Score = 41.6 bits (96), Expect(2) = 5e-23
Identities = 19/45 (42%), Positives = 29/45 (64%)
Frame = +2
Query: 1502 ISQVLVQWQGKTPEEATWEDLATIRSEFPVFNLEDKVVSSEGSIV 1546
I++VL+Q + EATWE + I+ +FP F+LEDKV G ++
Sbjct: 353 IAEVLIQLEDLPDFEATWESVEVIKEQFPSFHLEDKVTLLGGVLL 487
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,393,944
Number of Sequences: 63676
Number of extensions: 920141
Number of successful extensions: 4554
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 4464
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4531
length of query: 1596
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1486
effective length of database: 5,635,272
effective search space: 8374014192
effective search space used: 8374014192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0170.6


