

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0170.5
(603 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
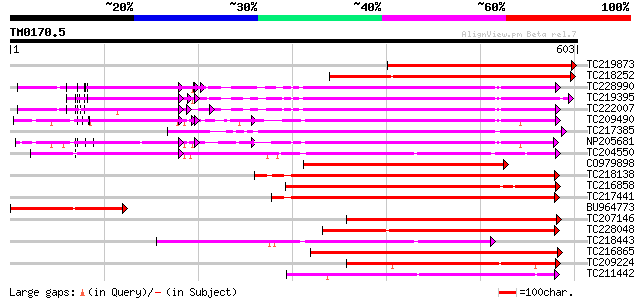
Score E
Sequences producing significant alignments: (bits) Value
TC219873 similar to UP|Q9FM23 (Q9FM23) Receptor-like protein kin... 372 e-103
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 349 2e-96
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 294 7e-80
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 292 3e-79
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 291 4e-79
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 272 4e-73
TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase P... 269 2e-72
NP205681 receptor protein kinase-like protein 258 5e-69
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 248 7e-66
CO979898 246 3e-65
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 238 6e-63
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 236 2e-62
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 234 8e-62
BU964773 homologue to GP|10177448|dbj receptor-like protein kina... 211 1e-54
TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, pa... 210 1e-54
TC228048 homologue to UP|Q8LA44 (Q8LA44) Receptor protein kinase... 210 2e-54
TC218443 similar to GB|BAB85647.1|19032343|AB076907 inflorescenc... 203 2e-52
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 202 5e-52
TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR1... 202 5e-52
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 201 6e-52
>TC219873 similar to UP|Q9FM23 (Q9FM23) Receptor-like protein kinase, partial
(32%)
Length = 864
Score = 372 bits (955), Expect = e-103
Identities = 184/203 (90%), Positives = 192/203 (93%), Gaps = 2/203 (0%)
Frame = +1
Query: 402 ENTEQP--LNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFG 459
ENT+Q LNWNDRL IALGSA+GLAYLHHEC PK+VH +IKSSNILL+ENMEPHISDFG
Sbjct: 1 ENTQQRQLLNWNDRLKIALGSAQGLAYLHHECSPKVVHCNIKSSNILLDENMEPHISDFG 180
Query: 460 LAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSF 519
LAKLLVDE AHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSF
Sbjct: 181 LAKLLVDEKAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSF 360
Query: 520 ANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQV 579
RGLNVVGWMNTL +ENRLEDVVD+RCTDADAGTLEVILELAARCTD NADDRPSMNQV
Sbjct: 361 VKRGLNVVGWMNTLLRENRLEDVVDKRCTDADAGTLEVILELAARCTDGNADDRPSMNQV 540
Query: 580 LQLLEQEVMSPCPSDFYESHSDH 602
LQLLEQEVMSPCPS+FYESHSDH
Sbjct: 541 LQLLEQEVMSPCPSEFYESHSDH 609
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 349 bits (896), Expect = 2e-96
Identities = 165/261 (63%), Positives = 214/261 (81%)
Frame = +1
Query: 341 AVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLL 400
A+KRI + EG D+ FERELEILGSIKH LVNLRGYC P+++LLIYDYL GSLD+ L
Sbjct: 1 ALKRIVKLNEGFDRFFERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEAL 180
Query: 401 HENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGL 460
HE +Q L+W+ RLNI +G+A+GLAYLHH+C P+I+HRDIKSSNILL+ N+E +SDFGL
Sbjct: 181 HERADQ-LDWDSRLNIIMGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLEARVSDFGL 357
Query: 461 AKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFA 520
AKLL DE++H+TT+VAGTFGYLAPEY+QSGRATEKSDVYSFGVL LE+++GKRPTD +F
Sbjct: 358 AKLLEDEESHITTIVAGTFGYLAPEYMQSGRATEKSDVYSFGVLTLEVLSGKRPTDAAFI 537
Query: 521 NRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVL 580
+G N+VGW+N L ENR ++VD C +L+ +L +A +C ++ +DRP+M++V+
Sbjct: 538 EKGPNIVGWLNFLITENRPREIVDPLCEGVQMESLDALLSVAIQCVSSSPEDRPTMHRVV 717
Query: 581 QLLEQEVMSPCPSDFYESHSD 601
QLLE EV++PCPSDFY+S+SD
Sbjct: 718 QLLESEVVTPCPSDFYDSNSD 780
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 294 bits (753), Expect = 7e-80
Identities = 180/510 (35%), Positives = 276/510 (53%), Gaps = 5/510 (0%)
Frame = +1
Query: 81 SQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIG 140
++ G I P IG L +L ++ N G I EI+ C L + L N G IP+ I
Sbjct: 1540 NEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKIT 1719
Query: 141 NLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGN 200
++ LN L+LS N G+IP ++ + L ++ S N FSG +P G F SF+GN
Sbjct: 1720 SMRILNYLNLSRNHLDGSIPGNIASMQSLTSVDFSYNNFSGLVPGTGQFGYFNYTSFLGN 1899
Query: 201 LDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVT 260
+LCG + PC+ PH K S +K++L+ + +L V
Sbjct: 1900 PELCGPYLG-PCKDGVANGPRQPHV---------KGPFSSSLKLLLVIGLLVCSILFAVA 2049
Query: 261 LSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIEKLESLEEEDI 320
F K RA+ + ++ + + KL F L +T ++ L+ L+E++I
Sbjct: 2050 AIF--------KARALKKASEAR-------AWKLTAFQR-LDFTVDDV---LDCLKEDNI 2172
Query: 321 VGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC--DQVFERELEILGSIKHINLVNLRGYC 378
+G GG G VY+ M + AVKR+ G D F E++ LG I+H ++V L G+C
Sbjct: 2173 IGKGGAGIVYKGAMPNGDNVAVKRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFC 2352
Query: 379 RLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHR 438
LL+Y+Y+ GSL ++LH L+W+ R IA+ +A+GL YLHH+C P IVHR
Sbjct: 2353 SNHETNLLVYEYMPNGSLGEVLHGKKGGHLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHR 2532
Query: 439 DIKSSNILLNENMEPHISDFGLAKLLVDEDA-HVTTVVAGTFGYLAPEYLQSGRATEKSD 497
D+KS+NILL+ N E H++DFGLAK L D A + +AG++GY+APEY + + EKSD
Sbjct: 2533 DVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLKVDEKSD 2712
Query: 498 VYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENR--LEDVVDRRCTDADAGTL 555
VYSFGV+LLELVTG++P F + G+++V W+ + N+ + V+D R +
Sbjct: 2713 VYSFGVVLLELVTGRKPVG-EFGD-GVDIVQWVRKMTDSNKEGVLKVLDSRLPSVPLHEV 2886
Query: 556 EVILELAARCTDANADDRPSMNQVLQLLEQ 585
+ +A C + A +RP+M +V+Q+L +
Sbjct: 2887 MHVFYVAMLCVEEQAVERPTMREVVQILTE 2976
Score = 93.6 bits (231), Expect = 2e-19
Identities = 62/195 (31%), Positives = 95/195 (47%), Gaps = 3/195 (1%)
Frame = +1
Query: 9 VFILIFTTVFTPSSLALTQ-DGLTLLEIKGALNDTKNVLSNWQEFDESP-CAWTGITCHP 66
V +L F + + + +++ L + +D + LS+W +P C+W G+TC
Sbjct: 106 VLVLFFLFLHSLQAARISEYRALLSFKASSLTDDPTHALSSWNS--STPFCSWFGLTC-- 273
Query: 67 GDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL 126
D + V S+NL L G +S + L L L+L N G IP + + LR L L
Sbjct: 274 -DSRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASFSALSALRFLNL 450
Query: 127 RANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PD 185
N F PS + L L +LDL +N+ G +P S+ +P L+ L+L NFFSG+I P+
Sbjct: 451 SNNVFNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPPE 630
Query: 186 IGVLSTFQKNSFIGN 200
G Q + GN
Sbjct: 631 YGTWQHLQYLALSGN 675
Score = 76.6 bits (187), Expect = 3e-14
Identities = 49/131 (37%), Positives = 68/131 (51%), Gaps = 2/131 (1%)
Frame = +1
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL-RANYF 131
+R ++L + G I P G LQ LAL N L G I E+ N + LR LY+ N +
Sbjct: 577 LRHLHLGGNFFSGQIPPEYGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIGYYNTY 756
Query: 132 QGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGVLS 190
GGIP +IGNL L LD + G IP+ LG+L +L L L N SG + P++G L
Sbjct: 757 SGGIPPEIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLK 936
Query: 191 TFQKNSFIGNL 201
+ + N+
Sbjct: 937 SLKSMDLSNNM 969
Score = 74.3 bits (181), Expect = 1e-13
Identities = 45/128 (35%), Positives = 66/128 (51%), Gaps = 6/128 (4%)
Frame = +1
Query: 80 YSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDI 139
Y+ G I P IG LS L RL L G IP E+ L L+L+ N G + ++
Sbjct: 745 YNTYSGGIPPEIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPEL 924
Query: 140 GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD-IGVLSTFQ----- 193
G+L L +DLS+N G +P+S L +L +LNL N G IP+ +G L +
Sbjct: 925 GSLKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLW 1104
Query: 194 KNSFIGNL 201
+N+F G++
Sbjct: 1105ENNFTGSI 1128
Score = 72.4 bits (176), Expect = 5e-13
Identities = 45/124 (36%), Positives = 63/124 (50%)
Frame = +1
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
GI G+ VR ++ Y L G I +GKL L L L N+L G + E+ +
Sbjct: 763 GIPPEIGNLSNLVR-LDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKS 939
Query: 121 LRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFS 180
L+++ L N G +P+ L L +L+L N GAIP +G LP L+VL L N F+
Sbjct: 940 LKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFT 1119
Query: 181 GEIP 184
G IP
Sbjct: 1120GSIP 1131
Score = 71.6 bits (174), Expect = 9e-13
Identities = 41/113 (36%), Positives = 58/113 (51%)
Frame = +1
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
++S++L + L G + S +L L L L +N LHG IP + L L L N F
Sbjct: 940 LKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFT 1119
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD 185
G IP ++GN L ++DLSSN G +P ++ LQ L N+ G IPD
Sbjct: 1120 GSIPQNLGNNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPD 1278
Score = 66.2 bits (160), Expect = 4e-11
Identities = 41/119 (34%), Positives = 60/119 (49%), Gaps = 1/119 (0%)
Frame = +1
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNL 142
L G I S+GK L R+ + +N L+G IP + +L + L+ N G P D
Sbjct: 1258 LFGPIPDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIA 1437
Query: 143 PFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGVLSTFQKNSFIGN 200
L + LS+N G++PS++G +Q L L+ N F+G I P IG+L K F N
Sbjct: 1438 TDLGQISLSNNQLSGSLPSTIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHN 1614
Score = 62.4 bits (150), Expect = 6e-10
Identities = 42/127 (33%), Positives = 61/127 (47%), Gaps = 1/127 (0%)
Frame = +1
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNL 142
L G I + L +L ++ L N L G P + + T+L + L N G +PS IGN
Sbjct: 1330 LNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLPSTIGNF 1509
Query: 143 PFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGVLSTFQKNSFIGNL 201
+ L L+ N F G IP +G L L ++ S N FSG I P+I K + +
Sbjct: 1510 TSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEI------SKCKLLTFI 1671
Query: 202 DLCGRQI 208
DL G ++
Sbjct: 1672 DLSGNEL 1692
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 292 bits (748), Expect = 3e-79
Identities = 189/532 (35%), Positives = 284/532 (52%), Gaps = 5/532 (0%)
Frame = +3
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
V+ + L + G I P IG+L +L ++ N G I EI+ C L L L N
Sbjct: 1200 VQKLILDGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELS 1379
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTF 192
G IP++I + LN L+LS N G IPSS+ + L ++ S N SG +P G S F
Sbjct: 1380 GDIPNEITGMRILNYLNLSRNHLVGGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYF 1559
Query: 193 QKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTT 252
SF+GN DLCG + C+ PH K SS + ++++G
Sbjct: 1560 NYTSFLGNPDLCGPYLGA-CKDGVANGAHQPHV---------KGLSSSFKLLLVVG---- 1697
Query: 253 LGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIEKL 312
LLL +++F A+ + +KK A KL F L +T ++ L
Sbjct: 1698 ---LLLCSIAFA--------VAAIFKARSLKKASGARA-WKLTAFQR-LDFTVDDV---L 1829
Query: 313 ESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC--DQVFERELEILGSIKHIN 370
L+E++I+G GG G VY+ M + AVKR+ G D F E++ LG I+H +
Sbjct: 1830 HCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSHDHGFNAEIQTLGRIRHRH 2009
Query: 371 LVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHE 430
+V L G+C LL+Y+Y+ GSL ++LH L+W+ R IA+ +A+GL YLHH+
Sbjct: 2010 IVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGHLHWDTRYKIAVEAAKGLCYLHHD 2189
Query: 431 CCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDE-DAHVTTVVAGTFGYLAPEYLQS 489
C P IVHRD+KS+NILL+ N E H++DFGLAK L D + + +AG++GY+APEY +
Sbjct: 2190 CSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAIAGSYGYIAPEYAYT 2369
Query: 490 GRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENR--LEDVVDRRC 547
+ EKSDVYSFGV+LLEL+TG++P F + G+++V W+ + N+ + V+D R
Sbjct: 2370 LKVDEKSDVYSFGVVLLELITGRKPVG-EFGD-GVDIVQWVRKMTDSNKEGVLKVLDPRL 2543
Query: 548 TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSDFYESH 599
+ + +A C + A +RP+M +V+Q+L + P P D E +
Sbjct: 2544 PSVPLHEVMHVFYVAMLCVEEQAVERPTMREVVQILTE---LPKPPDSKEGN 2690
Score = 84.0 bits (206), Expect = 2e-16
Identities = 52/133 (39%), Positives = 72/133 (54%), Gaps = 2/133 (1%)
Frame = +3
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL-RAN 129
Q +R ++L + G I P G+ RLQ LA+ N L G IP EI N + LR LY+ N
Sbjct: 255 QNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELEGTIPPEIGNLSSLRELYIGYYN 434
Query: 130 YFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGV 188
+ GGIP +IGNL L LD + G IP++LG+L L L L N SG + P++G
Sbjct: 435 TYTGGIPPEIGNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGN 614
Query: 189 LSTFQKNSFIGNL 201
L + + N+
Sbjct: 615 LKSLKSMDLSNNM 653
Score = 77.4 bits (189), Expect = 2e-14
Identities = 46/125 (36%), Positives = 68/125 (53%)
Frame = +3
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
GI G+ + VR ++ Y L G I ++GKL +L L L N+L G + E+ N
Sbjct: 447 GIPPEIGNLSELVR-LDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKS 623
Query: 121 LRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFS 180
L+++ L N G IP+ G L + +L+L N GAIP +G LP L+V+ L N F+
Sbjct: 624 LKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFT 803
Query: 181 GEIPD 185
G IP+
Sbjct: 804 GSIPE 818
Score = 73.9 bits (180), Expect = 2e-13
Identities = 46/128 (35%), Positives = 66/128 (50%), Gaps = 6/128 (4%)
Frame = +3
Query: 80 YSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDI 139
Y+ G I P IG LS L RL L G IP + +L L+L+ N G + ++
Sbjct: 429 YNTYTGGIPPEIGNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPEL 608
Query: 140 GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD-IGVLSTFQ----- 193
GNL L +DLS+N G IP+ G L ++ +LNL N G IP+ IG L +
Sbjct: 609 GNLKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLW 788
Query: 194 KNSFIGNL 201
+N+F G++
Sbjct: 789 ENNFTGSI 812
Score = 69.7 bits (169), Expect = 4e-12
Identities = 42/113 (37%), Positives = 57/113 (50%)
Frame = +3
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
++S++L + L G I G+L + L L +N LHG IP I L + L N F
Sbjct: 624 LKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFT 803
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD 185
G IP +G LN++DLSSN G +P+ L LQ L NF G IP+
Sbjct: 804 GSIPEGLGKNGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPE 962
Score = 68.9 bits (167), Expect = 6e-12
Identities = 37/102 (36%), Positives = 57/102 (55%)
Frame = +3
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNL 142
L G +S + L L L+L N G IP ++ + LR L L N F PS++ L
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRL 182
Query: 143 PFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
L +LDL +N+ G +P ++ ++ +L+ L+L NFFSG+IP
Sbjct: 183 QNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIP 308
Score = 62.8 bits (151), Expect = 4e-10
Identities = 41/121 (33%), Positives = 60/121 (48%), Gaps = 1/121 (0%)
Frame = +3
Query: 75 SINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGG 134
+++L ++ G I PS+ LS L+ L L N + P+E++ L L L N G
Sbjct: 51 NLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRLQNLEVLDLYNNNMTGV 230
Query: 135 IPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGVLSTFQ 193
+P + + L L L N F G IP GR LQ L +S N G I P+IG LS+ +
Sbjct: 231 LPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELEGTIPPEIGNLSSLR 410
Query: 194 K 194
+
Sbjct: 411 E 413
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 291 bits (746), Expect = 4e-79
Identities = 191/539 (35%), Positives = 283/539 (52%), Gaps = 29/539 (5%)
Frame = +3
Query: 76 INLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPN---------------------- 113
I L +QL G +SPSIG S +Q+L L N G IP
Sbjct: 1386 ITLSNNQLSGALSPSIGNFSSVQKLLLDGNMFTGRIPTQIGRLQQLSKIDFSGNKFSGPI 1565
Query: 114 --EITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQV 171
EI+ C L L L N G IP++I + LN L+LS N G+IPSS+ + L
Sbjct: 1566 APEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSKNHLVGSIPSSISSMQSLTS 1745
Query: 172 LNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAA 231
++ S N SG +P G S F SF+GN DLCG + C+ PH
Sbjct: 1746 VDFSYNNLSGLVPGTGQFSYFNYTSFLGNPDLCGPYLGA-CKGGVANGAHQPHV------ 1904
Query: 232 VPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEAS 291
K SS ++++G LLL +++F A+ + +KK + A
Sbjct: 1905 ---KGLSSSLKLLLVVG-------LLLCSIAFA--------VAAIFKARSLKKASEARA- 2027
Query: 292 TKLITFHGDLPYTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREG 351
KL F L +T ++ L L+E++I+G GG G VY+ M + AVKR+ G
Sbjct: 2028 WKLTAFQR-LDFTVDDV---LHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRG 2195
Query: 352 C--DQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLN 409
D F E++ LG I+H ++V L G+C LL+Y+Y+ GSL ++LH L+
Sbjct: 2196 SSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGHLH 2375
Query: 410 WNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDE-D 468
W+ R IA+ +A+GL YLHH+C P IVHRD+KS+NILL+ N E H++DFGLAK L D
Sbjct: 2376 WDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGT 2555
Query: 469 AHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVG 528
+ + +AG++GY+APEY + + EKSDVYSFGV+LLEL+TG++P F + G+++V
Sbjct: 2556 SECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVG-EFGD-GVDIVQ 2729
Query: 529 WMNTLQKENR--LEDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQ 585
W+ + N+ + V+D R + + +A C + A +RP+M +V+Q+L +
Sbjct: 2730 WVRKMTDSNKEGVLKVLDPRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVVQILTE 2906
Score = 94.7 bits (234), Expect = 1e-19
Identities = 59/178 (33%), Positives = 93/178 (52%), Gaps = 2/178 (1%)
Frame = +3
Query: 9 VFILIFTTVFTPSSL-ALTQDGLTLLEIKGALND-TKNVLSNWQEFDESPCAWTGITCHP 66
+F+ +F P +L A + LL ++ + D T VLS+W C+W G+TC
Sbjct: 36 LFVFLFFHFHFPETLSAPISEYRALLSLRSVITDATPPVLSSWNA-SIPYCSWLGVTC-- 206
Query: 67 GDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL 126
D + V ++NL L G +S + L L L+L N G IP ++ + LR L L
Sbjct: 207 -DNRRHVTALNLTGLDLSGTLSADVAHLPFLSNLSLAANKFSGPIPPSLSALSGLRYLNL 383
Query: 127 RANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
N F PS++ L L +LDL +N+ G +P ++ ++ +L+ L+L NFFSG+IP
Sbjct: 384 SNNVFNETFPSELWRLQSLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIP 557
Score = 87.4 bits (215), Expect = 2e-17
Identities = 57/149 (38%), Positives = 80/149 (53%), Gaps = 2/149 (1%)
Frame = +3
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL-RAN 129
Q +R ++L + G I P G+ RLQ LA+ N L G IP EI N T LR LY+ N
Sbjct: 504 QNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELDGTIPPEIGNLTSLRELYIGYYN 683
Query: 130 YFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEI-PDIGV 188
+ GGIP +IGNL L LD++ + G IP++LG+L L L L N SG + P++G
Sbjct: 684 TYTGGIPPEIGNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGN 863
Query: 189 LSTFQKNSFIGNLDLCGRQIQKPCRTSFG 217
L + + ++DL + SFG
Sbjct: 864 LKSLK------SMDLSNNMLSGEIPASFG 932
Score = 77.0 bits (188), Expect = 2e-14
Identities = 45/125 (36%), Positives = 68/125 (54%)
Frame = +3
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
GI G+ + VR +++ Y L G I ++GKL +L L L N+L G + E+ N
Sbjct: 696 GIPPEIGNLSELVR-LDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKS 872
Query: 121 LRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFS 180
L+++ L N G IP+ G L + +L+L N GAIP +G LP L+V+ L N +
Sbjct: 873 LKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLT 1052
Query: 181 GEIPD 185
G IP+
Sbjct: 1053GSIPE 1067
Score = 74.7 bits (182), Expect = 1e-13
Identities = 44/115 (38%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Frame = +3
Query: 80 YSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDI 139
Y+ G I P IG LS L RL + +L G IP + +L L+L+ N G + ++
Sbjct: 678 YNTYTGGIPPEIGNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPEL 857
Query: 140 GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD-IGVLSTFQ 193
GNL L +DLS+N G IP+S G L ++ +LNL N G IP+ IG L +
Sbjct: 858 GNLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALE 1022
Score = 68.6 bits (166), Expect = 8e-12
Identities = 42/113 (37%), Positives = 56/113 (49%)
Frame = +3
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
++S++L + L G I S G+L + L L +N LHG IP I L + L N
Sbjct: 873 LKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLT 1052
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD 185
G IP +G LN++DLSSN G +P L LQ L NF G IP+
Sbjct: 1053GSIPEGLGKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPE 1211
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 272 bits (695), Expect = 4e-73
Identities = 179/526 (34%), Positives = 279/526 (53%), Gaps = 13/526 (2%)
Frame = +1
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+++++L ++ G I + L L + + N+L G IP +T C L A+ L N +
Sbjct: 1540 LQTLSLDANEFVGEIPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVDLSRNMLE 1719
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTF 192
G IP I NL L+I ++S N G +P + + L L+LS N F G++P G + F
Sbjct: 1720 GKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPTGGQFAVF 1899
Query: 193 QKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMK----VVLIG 248
+ SF GN +LC C S +P D+A KR +K +V++
Sbjct: 1900 SEKSFAGNPNLC---TSHSCPNSSLYP--------DDAL--KKRRGPWSLKSTRVIVIVI 2040
Query: 249 AMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEI 308
A+ T LL+ VT+ + R ++ A T +T L + + ++
Sbjct: 2041 ALGTAALLVAVTVYMMRRRKMNL------------------AKTWKLTAFQRLNFKAEDV 2166
Query: 309 IEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC-DQVFERELEILGSIK 367
+E L+EE+I+G GG G VYR M + A+KR+ + G D F+ E+E LG I+
Sbjct: 2167 VE---CLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGAGSGRNDYGFKAEIETLGKIR 2337
Query: 368 HINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYL 427
H N++ L GY LL+Y+Y+ GSL + LH L W R IA+ +A+GL YL
Sbjct: 2338 HRNIMRLLGYVSNKETNLLLYEYMPNGSLGEWLHGAKGGHLKWEMRYKIAVEAAKGLCYL 2517
Query: 428 HHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVT-TVVAGTFGYLAPEY 486
HH+C P I+HRD+KS+NILL+ ++E H++DFGLAK L D A + + +AG++GY+APEY
Sbjct: 2518 HHDCSPLIIHRDVKSNNILLDGDLEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEY 2697
Query: 487 LQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLED----- 541
+ + EKSDVYSFGV+LLEL+ G++P F + G+++VGW+N + E
Sbjct: 2698 AYTLKVDEKSDVYSFGVVLLELIIGRKPVG-EFGD-GVDIVGWVNKTRLELAQPSDAALV 2871
Query: 542 --VVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQ 585
VVD R + ++ + +A C RP+M +V+ +L +
Sbjct: 2872 LAVVDPRLSGYPLTSVIYMFNIAMMCVKEMGPARPTMREVVHMLSE 3009
Score = 96.3 bits (238), Expect = 3e-20
Identities = 66/189 (34%), Positives = 104/189 (54%), Gaps = 8/189 (4%)
Frame = +1
Query: 5 FPIWVFIL-IFTTVFTPSSLALTQDGLTLLEIKGALNDTK---NVLSNWQEFDE--SPCA 58
+ + +FI I+ V T SS D +LL++K ++ K + L +W+ F + C
Sbjct: 109 YTLLLFIFFIWLRVATCSSFT---DMESLLKLKDSMKGDKAKDDALHDWKFFPSLSAHCF 279
Query: 59 WTGITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNC 118
++G+ C D E RV +IN+ + L G + P IG+L +L+ L + QN+L G++P E+
Sbjct: 280 FSGVKC---DRELRVVAINVSFVPLFGHLPPEIGQLDKLENLTVSQNNLTGVLPKELAAL 450
Query: 119 TELRALYLRANYFQGGIPSDIGNLPF--LNILDLSSNSFKGAIPSSLGRLPHLQVLNLST 176
T L+ L + N F G P I LP L +LD+ N+F G +P L +L L+ L L
Sbjct: 451 TSLKHLNISHNVFSGHFPGQI-ILPMTKLEVLDVYDNNFTGPLPVELVKLEKLKYLKLDG 627
Query: 177 NFFSGEIPD 185
N+FSG IP+
Sbjct: 628 NYFSGSIPE 654
Score = 79.3 bits (194), Expect = 4e-15
Identities = 61/199 (30%), Positives = 99/199 (49%), Gaps = 9/199 (4%)
Frame = +1
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+R ++L L G I PS+ L+ L L L N+L G IP+E++ L +L L N
Sbjct: 823 LRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGTIPSELSAMVSLMSLDLSINDLT 1002
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP----DIGV 188
G IP L L +++ N+ +G++PS +G LP+L+ L L N FS +P G
Sbjct: 1003 GEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLETLQLWDNNFSFVLPPNLGQNGK 1182
Query: 189 LSTFQ--KNSFIGNL--DLC-GRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMK 243
L F KN F G + DLC ++Q T F IP+ + ++ R+S++Y+
Sbjct: 1183 LKFFDVIKNHFTGLIPRDLCKSGRLQTIMITDNFFRGPIPNEIGNCKSLTKIRASNNYLN 1362
Query: 244 VVLIGAMTTLGLLLLVTLS 262
V+ + L + ++ L+
Sbjct: 1363 GVVPSGIFKLPSVTIIELA 1419
Score = 76.6 bits (187), Expect = 3e-14
Identities = 45/123 (36%), Positives = 64/123 (51%), Gaps = 6/123 (4%)
Frame = +1
Query: 85 GIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPF 144
G+I + K RLQ + + N G IPNEI NC L + NY G +PS I LP
Sbjct: 1219 GLIPRDLCKSGRLQTIMITDNFFRGPIPNEIGNCKSLTKIRASNNYLNGVVPSGIFKLPS 1398
Query: 145 LNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP----DIGVLST--FQKNSFI 198
+ I++L++N F G +P + L +L LS N FSG+IP ++ L T N F+
Sbjct: 1399 VTIIELANNRFNGELPPEISG-ESLGILTLSNNLFSGKIPPALKNLRALQTLSLDANEFV 1575
Query: 199 GNL 201
G +
Sbjct: 1576 GEI 1584
Score = 73.6 bits (179), Expect = 2e-13
Identities = 36/122 (29%), Positives = 69/122 (56%), Gaps = 6/122 (4%)
Frame = +1
Query: 86 IISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFL 145
++ P++G+ +L+ + +N G+IP ++ L+ + + N+F+G IP++IGN L
Sbjct: 1150 VLPPNLGQNGKLKFFDVIKNHFTGLIPRDLCKSGRLQTIMITDNFFRGPIPNEIGNCKSL 1329
Query: 146 NILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP------DIGVLSTFQKNSFIG 199
+ S+N G +PS + +LP + ++ L+ N F+GE+P +G+L T N F G
Sbjct: 1330 TKIRASNNYLNGVVPSGIFKLPSVTIIELANNRFNGELPPEISGESLGIL-TLSNNLFSG 1506
Query: 200 NL 201
+
Sbjct: 1507 KI 1512
Score = 67.4 bits (163), Expect = 2e-11
Identities = 47/135 (34%), Positives = 66/135 (48%), Gaps = 14/135 (10%)
Frame = +1
Query: 78 LPYSQLG------GIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYF 131
L Y +LG G I P G + L+ L L +L G IP + N T L L+L+ N
Sbjct: 748 LRYLKLGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNL 927
Query: 132 QGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTN-------FFSGEIP 184
G IPS++ + L LDLS N G IP S +L +L ++N N F GE+P
Sbjct: 928 TGTIPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELP 1107
Query: 185 DIGVLSTFQKN-SFI 198
++ L + N SF+
Sbjct: 1108NLETLQLWDNNFSFV 1152
>TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase PERK1-like,
partial (80%)
Length = 1604
Score = 269 bits (688), Expect = 2e-72
Identities = 165/429 (38%), Positives = 242/429 (55%), Gaps = 5/429 (1%)
Frame = +1
Query: 169 LQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESD 228
L +LN+S N G IP + F +SFIGN LCG + PC
Sbjct: 7 LSLLNVSYNKLFGVIPTSNNFTRFPPDSFIGNPGLCGNWLNLPCH--------------- 141
Query: 229 EAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDP 288
A P++R + K ++G TLG L+++ L + + + + + + D
Sbjct: 142 -GARPSERVT--LSKAAILGI--TLGALVIL----LMVLVAACRPHSPSPFPDGSFDKPI 294
Query: 289 EAST-KLITFHGDLP-YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRID 346
S KL+ H ++ + +I+ E+L E+ I+G G TVY+ V+ +C A+KRI
Sbjct: 295 NFSPPKLVILHMNMALHVYEDIMRMTENLSEKYIIGYGASSTVYKCVLKNCKPVAIKRIY 474
Query: 347 RSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQ 406
C + FE ELE +GSIKH NLV+L+GY P LL YDY+ GSL DLLH T++
Sbjct: 475 SHYPQCIKEFETELETVGSIKHRNLVSLQGYSLSPYGHLLFYDYMENGSLWDLLHGPTKK 654
Query: 407 P-LNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLV 465
L+W RL IALG+A+GLAYLHH+CCP+I+HRD+KSSNILL+ + EPH++DFG+AK L
Sbjct: 655 KKLDWELRLKIALGAAQGLAYLHHDCCPRIIHRDVKSSNILLDADFEPHLTDFGIAKSLC 834
Query: 466 DEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLN 525
+H +T + GT GY+ PEY ++ TEKSDVYS+G++LLEL+TG++ D N
Sbjct: 835 PSKSHTSTYIMGTIGYIDPEYARTSHLTEKSDVYSYGIVLLELLTGRKAVD-----NESN 999
Query: 526 VVGWMNTLQKENRLEDVVDR--RCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLL 583
+ + + N + + VD T D G ++ + +LA CT DRP+M++V ++L
Sbjct: 1000LHHLILSKAATNAVMETVDPDITATCKDLGAVKKVYQLALLCTKRQPADRPTMHEVTRVL 1179
Query: 584 EQEVMSPCP 592
V S P
Sbjct: 1180GSLVPSSIP 1206
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 258 bits (659), Expect = 5e-69
Identities = 171/520 (32%), Positives = 270/520 (51%), Gaps = 9/520 (1%)
Frame = +1
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+++++L ++ G I + L L + + N+L G IP T C L A+ L N
Sbjct: 1450 LQTLSLDTNEFLGEIPGEVFDLPMLTVVNISGNNLTGPIPTTFTRCVSLAAVDLSRNMLD 1629
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTF 192
G IP + NL L+I ++S N G++P + + L L+LS N F G++P G F
Sbjct: 1630 GEIPKGMKNLTDLSIFNVSINQISGSVPDEIRFMLSLTTLDLSYNNFIGKVPTGGQFLVF 1809
Query: 193 QKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTT 252
SF GN +LC P++ + P S+ +V++ A+ T
Sbjct: 1810 SDKSFAGNPNLCSSHS-------------CPNSSLKKRRGPWSLKSTRV--IVMVIALAT 1944
Query: 253 LGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIEKL 312
+L+ T Y ++++ + KL F L + E++E
Sbjct: 1945 AAILVAGT-----------------EYMRRRRKLKLAMTWKLTGFQR-LNLKAEEVVE-- 2064
Query: 313 ESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC-DQVFERELEILGSIKHINL 371
L+EE+I+G GG G VYR M + A+KR+ + G D F+ E+E +G I+H N+
Sbjct: 2065 -CLKEENIIGKGGAGIVYRGSMRNGSDVAIKRLVGAGSGRNDYGFKAEIETVGKIRHRNI 2241
Query: 372 VNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHEC 431
+ L GY LL+Y+Y+ GSL + LH L W R IA+ +A+GL YLHH+C
Sbjct: 2242 MRLLGYVSNKETNLLLYEYMPNGSLGEWLHGAKGGHLKWEMRYKIAVEAAKGLCYLHHDC 2421
Query: 432 CPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVD-EDAHVTTVVAGTFGYLAPEYLQSG 490
P I+HRD+KS+NILL+ + E H++DFGLAK L D + + +AG++GY+APEY +
Sbjct: 2422 SPLIIHRDVKSNNILLDAHFEAHVADFGLAKFLYDLGSSQSMSSIAGSYGYIAPEYAYTL 2601
Query: 491 RATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKE-NRLED------VV 543
+ EKSDVYSFGV+LLEL+ G++P F + G+++VGW+N + E ++ D VV
Sbjct: 2602 KVDEKSDVYSFGVVLLELIIGRKPVG-EFGD-GVDIVGWVNKTRLELSQPSDAAVVLAVV 2775
Query: 544 DRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLL 583
D R + ++ + +A C RP+M +V+ +L
Sbjct: 2776 DPRLSGYPLISVIYMFNIAMMCVKEVGPTRPTMREVVHML 2895
Score = 89.7 bits (221), Expect = 3e-18
Identities = 65/187 (34%), Positives = 103/187 (54%), Gaps = 8/187 (4%)
Frame = +1
Query: 7 IWVFILIFTTVFTPSSLALTQDGLTLLEIKGALNDTK---NVLSNWQEFDES---PCAWT 60
++VF I+ V T SS + D LL++K ++ + + L +W+ F S C ++
Sbjct: 31 LFVFF-IWLHVATCSSFS---DMDALLKLKESMKGDRAKDDALHDWK-FSTSLSAHCFFS 195
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
G++C D E RV +IN+ + L G + P IG+L +L+ L + QN+L G +P E+ T
Sbjct: 196 GVSC---DQELRVVAINVSFVPLFGHVPPEIGELDKLENLTISQNNLTGELPKELAALTS 366
Query: 121 LRALYLRANYFQGGIPSDIGNLPF--LNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNF 178
L+ L + N F G P I LP L +LD+ N+F G++P +L L+ L L N+
Sbjct: 367 LKHLNISHNVFSGYFPGKI-ILPMTELEVLDVYDNNFTGSLPEEFVKLEKLKYLKLDGNY 543
Query: 179 FSGEIPD 185
FSG IP+
Sbjct: 544 FSGSIPE 564
Score = 81.3 bits (199), Expect = 1e-15
Identities = 58/201 (28%), Positives = 104/201 (50%), Gaps = 9/201 (4%)
Frame = +1
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
+ ++ ++L L G I PS+ + L L L N+L G IP+E+++ L +L L N
Sbjct: 727 ESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGTIPSELSDMVSLMSLDLSFNG 906
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP-DIGVL 189
G IP+ L L +++ N+ +G++PS +G LP+L+ L L N FS E+P ++G
Sbjct: 907 LTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLETLQLWENNFSSELPQNLGQN 1086
Query: 190 STFQ-----KNSFIGNL--DLC-GRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHY 241
F+ KN F G + DLC ++Q T F IP+ ++ ++ R+S++Y
Sbjct: 1087GKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNEIANCKSLTKIRASNNY 1266
Query: 242 MKVVLIGAMTTLGLLLLVTLS 262
+ + + L + ++ L+
Sbjct: 1267LNGAVPSGIFKLPSVTIIELA 1329
Score = 79.7 bits (195), Expect = 3e-15
Identities = 45/127 (35%), Positives = 65/127 (50%), Gaps = 6/127 (4%)
Frame = +1
Query: 81 SQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIG 140
+ G+I + K RLQ + N HG IPNEI NC L + NY G +PS I
Sbjct: 1117 NHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNEIANCKSLTKIRASNNYLNGAVPSGIF 1296
Query: 141 NLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP----DIGVLST--FQK 194
LP + I++L++N F G +P + L +L LS N F+G+IP ++ L T
Sbjct: 1297 KLPSVTIIELANNRFNGELPPEISG-DSLGILTLSNNLFTGKIPPALKNLRALQTLSLDT 1473
Query: 195 NSFIGNL 201
N F+G +
Sbjct: 1474 NEFLGEI 1494
Score = 67.0 bits (162), Expect = 2e-11
Identities = 34/118 (28%), Positives = 63/118 (52%), Gaps = 6/118 (5%)
Frame = +1
Query: 90 SIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILD 149
++G+ + + + +N G+IP ++ L+ + N+F G IP++I N L +
Sbjct: 1072 NLGQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNEIANCKSLTKIR 1251
Query: 150 LSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP------DIGVLSTFQKNSFIGNL 201
S+N GA+PS + +LP + ++ L+ N F+GE+P +G+L T N F G +
Sbjct: 1252 ASNNYLNGAVPSGIFKLPSVTIIELANNRFNGELPPEISGDSLGIL-TLSNNLFTGKI 1422
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 248 bits (632), Expect = 7e-66
Identities = 183/563 (32%), Positives = 277/563 (48%), Gaps = 48/563 (8%)
Frame = +2
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
+ + I+L +++ G I IG LSRL+ L + N+L+G +P ++N + L L N
Sbjct: 956 RELNEISLSHNKFSGAIPNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNL 1135
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP------ 184
IP +G L L++L LS N F G IPSS+ + L+ L+LS N FSGEIP
Sbjct: 1136 LDNQIPQSLGRLRNLSVLILSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQ 1315
Query: 185 --------DIGVLS---------TFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAES 227
LS F +SF+GN+ LCG PC + VI
Sbjct: 1316 RSLNLFNVSYNSLSGSVPPLLAKKFNSSSFVGNIQLCGYSPSTPCFSQAPSQGVIAPPPE 1495
Query: 228 DEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKK--------ERAVMRY 279
++ S+ + +V+ G + + ++L L F IR S E + Y
Sbjct: 1496 VSKHHHHRKLSTKDIILVVAGVLLVVLIILCCVLLFCLIRKRSTSKAGNGQATEGRAVTY 1675
Query: 280 TDVK-----------KQVDPEASTKLITFHGDLPYTSSEIIEKLESLEEEDIVGSGGFGT 328
D K KQV L+ F G + +T+ +++ +I+G GT
Sbjct: 1676 EDRKRSPSSCWWLMLKQVG-RLEGNLVHFDGPMAFTADDLL-----CATAEIMGKSTNGT 1837
Query: 329 VYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSA-RLLI 387
VY+ ++ D AVKR+ + FE E+ +LG I+H N++ LR Y P +LL+
Sbjct: 1838 VYKAILEDGSQVAVKRLREKIAKGHREFESEVSVLGKIRHPNVLALRAYYLGPKGEKLLV 2017
Query: 388 YDYLAIGSLDDLLHEN-TEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNIL 446
+DY++ GSL LH TE ++W R+ IA ARGL LH + I+H ++ SSN+L
Sbjct: 2018 FDYMSKGSLASFLHGGGTETFIDWPTRMKIAQDLARGLFCLHSQ--ENIIHGNLTSSNVL 2191
Query: 447 LNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLL 506
L+EN I+DFGL++L+ AG GY APE + +A K+D+YS GV+LL
Sbjct: 2192 LDENTNAKIADFGLSRLMSTTANSNVIATAGALGYRAPELSKLKKANTKTDIYSLGVILL 2371
Query: 507 ELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTL--EVI--LELA 562
EL+T K P + GL++ W+ ++ KE +V D DA T+ E++ L+LA
Sbjct: 2372 ELLTRK---SPGVSMNGLDLPQWVASVVKEEWTNEVFDADLM-RDASTVGDELLNTLKLA 2539
Query: 563 ARCTDANADDRPSMNQVLQLLEQ 585
C D + RP ++QVLQ LE+
Sbjct: 2540 LHCVDPSPSARPEVHQVLQQLEE 2608
Score = 98.6 bits (244), Expect = 7e-21
Identities = 64/165 (38%), Positives = 84/165 (50%), Gaps = 3/165 (1%)
Frame = +2
Query: 23 LALTQDGLTLLE-IKGALNDTKNVLSNWQEFDESPCA--WTGITCHPGDGEQRVRSINLP 79
+ +T L LE K L D + L +W + C+ W GI C G +V I LP
Sbjct: 296 VVVTASNLLALEAFKQELADPEGFLRSWNDSGYGACSGGWVGIKCAQG----QVIVIQLP 463
Query: 80 YSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDI 139
+ L G I+ IG+L L++L+LH N + G IP+ + LR + L N G IP +
Sbjct: 464 WKGLRGRITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSL 643
Query: 140 GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G P L LDLS+N GAIP SL L LNLS N FSG +P
Sbjct: 644 GFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLP 778
>CO979898
Length = 869
Score = 246 bits (627), Expect = 3e-65
Identities = 118/219 (53%), Positives = 156/219 (70%), Gaps = 1/219 (0%)
Frame = -1
Query: 313 ESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLV 372
+ L +DI+GSGG+G VY + ++D A+KR++R D+ FERELE + IKH N+V
Sbjct: 869 QKLNSKDIIGSGGYGVVYELKLDDSTALAIKRLNRGTAERDKGFERELEAMADIKHRNIV 690
Query: 373 NLRGYCRLPSARLLIYDYLAIGSLDDLLH-ENTEQPLNWNDRLNIALGSARGLAYLHHEC 431
L GY P LLIY+ + GSLD LH + E+ L+W R IA G+ARG++YLHH+C
Sbjct: 689 TLHGYYTAPLYNLLIYELMPHGSLDSFLHGRSREKVLDWPTRYRIAAGAARGISYLHHDC 510
Query: 432 CPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGR 491
P I+HRDIKSSNILL++NM+ +SDFGLA L+ HV+T+VAGTFGYLAPEY +GR
Sbjct: 509 IPHIIHRDIKSSNILLDQNMDARVSDFGLATLMQPNKTHVSTIVAGTFGYLAPEYFDTGR 330
Query: 492 ATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWM 530
AT K DVYSFGV+LLEL+TGK+P+D +F G +V W+
Sbjct: 329 ATLKGDVYSFGVVLLELLTGKKPSDEAFMEEGTMLVTWV 213
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 238 bits (607), Expect = 6e-63
Identities = 135/331 (40%), Positives = 205/331 (61%), Gaps = 7/331 (2%)
Frame = +1
Query: 261 LSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-YTSSEIIEKLESLEEED 319
++F W R +E + DV + DPE G L ++ E+ +S ++
Sbjct: 22 IAFAWWRRRKPQEF----FFDVPAEEDPEVHL------GQLKRFSLRELQVATDSFSNKN 171
Query: 320 IVGSGGFGTVYRMVMNDCGTFAVKRIDRSRE-GCDQVFERELEILGSIKHINLVNLRGYC 378
I+G GGFG VY+ + D AVKR+ R G + F+ E+E++ H NL+ LRG+C
Sbjct: 172 ILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLLRLRGFC 351
Query: 379 RLPSARLLIYDYLAIGSLDDLLHENT--EQPLNWNDRLNIALGSARGLAYLHHECCPKIV 436
P+ RLL+Y Y+A GS+ L E ++PL+W R +ALGSARGL+YLH C PKI+
Sbjct: 352 MTPTERLLVYPYMANGSVASCLRERPPYQEPLDWPTRKRVALGSARGLSYLHDHCDPKII 531
Query: 437 HRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKS 496
HRD+K++NILL+E E + DFGLAKL+ +D HVTT V GT G++APEYL +G+++EK+
Sbjct: 532 HRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKT 711
Query: 497 DVYSFGVLLLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKENRLEDVVDRRC-TDADAG 553
DV+ +G++LLEL+TG+R D AN + ++ W+ L KE +LE +VD T+
Sbjct: 712 DVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEMLVDPDLQTNYIET 891
Query: 554 TLEVILELAARCTDANADDRPSMNQVLQLLE 584
+E ++++A CT + DRP M++V+++LE
Sbjct: 892 EVEQLIQVALLCTQGSPMDRPKMSEVVRMLE 984
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 236 bits (602), Expect = 2e-62
Identities = 128/300 (42%), Positives = 191/300 (63%), Gaps = 8/300 (2%)
Frame = +1
Query: 294 LITFHGDL-PYTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC 352
L TF L T +++++ + ++GSGGFG VY+ + D A+K++
Sbjct: 16 LATFEKPLRKLTFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQG 195
Query: 353 DQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQ--PLNW 410
D+ F E+E +G IKH NLV L GYC++ RLL+Y+Y+ GSL+D+LH+ + LNW
Sbjct: 196 DREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNW 375
Query: 411 NDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAH 470
R IA+G+ARGLA+LHH C P I+HRD+KSSN+LL+EN+E +SDFG+A+L+ D H
Sbjct: 376 AIRRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTH 555
Query: 471 VT-TVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPS-FANRGLNVVG 528
++ + +AGT GY+ PEY QS R + K DVYS+GV+LLEL+TGKRPTD + F + N+VG
Sbjct: 556 LSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDN--NLVG 729
Query: 529 WMNTLQKENRLEDVVDRRCTDADAG-TLEVI--LELAARCTDANADDRPSMNQVLQLLEQ 585
W+ K ++ D+ D D +E++ L++A C D RP+M QV+ + ++
Sbjct: 730 WVKQHAK-LKISDIFDPELMKEDPNLEMELLQHLKIAVSCLDDRPWRRPTMIQVMAMFKE 906
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 234 bits (597), Expect = 8e-62
Identities = 131/313 (41%), Positives = 196/313 (61%), Gaps = 7/313 (2%)
Frame = +2
Query: 279 YTDVKKQVDPEASTKLITFHGDLP-YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDC 337
+ DV + DPE G L ++ E+ ++ + I+G GGFG VY+ + D
Sbjct: 62 FFDVPAEEDPEVHL------GQLKRFSLRELQVATDNFSNKHILGRGGFGKVYKGRLADG 223
Query: 338 GTFAVKRIDRSR-EGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSL 396
AVKR+ R +G F+ E+E++ H NL+ LRG+C P+ RLL+Y Y+A GS+
Sbjct: 224 SLVAVKRLKEERTQGG*LQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSV 403
Query: 397 DDLLHENTEQ--PLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPH 454
L E E PL W +R IALGSARGLAYLH C PKI+HRD+K++NILL+E E
Sbjct: 404 ASCLRERQESQPPLGWPERKRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAV 583
Query: 455 ISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRP 514
+ DFGLAKL+ +D HVTT V GT G++APEYL +G+++EK+DV+ +GV+LLEL+TG+R
Sbjct: 584 VGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRA 763
Query: 515 TD-PSFAN-RGLNVVGWMNTLQKENRLEDVVDRRCTDA-DAGTLEVILELAARCTDANAD 571
D AN + ++ W+ L K+ +LE +VD + + +E ++++A CT +
Sbjct: 764 FDLARLANDDDVMLLDWVKGLLKDRKLETLVDADLQGSYNDEEVEQLIQVALLCTQGSPM 943
Query: 572 DRPSMNQVLQLLE 584
+RP M++V+++LE
Sbjct: 944 ERPKMSEVVRMLE 982
>BU964773 homologue to GP|10177448|dbj receptor-like protein kinase
{Arabidopsis thaliana}, partial (9%)
Length = 444
Score = 211 bits (536), Expect = 1e-54
Identities = 103/125 (82%), Positives = 110/125 (87%)
Frame = +1
Query: 1 MEKGFPIWVFILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWT 60
ME G W+ ++I TVF PSSLALT DG+TLLEIK LNDTKNVLSNWQ+FDES CAWT
Sbjct: 73 MEMGTVAWISLVIIVTVFCPSSLALTLDGMTLLEIKSTLNDTKNVLSNWQQFDESHCAWT 252
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
GI+CHPGD EQRVRSINLPY QLGGIISPSIGKLSRLQRLALHQNSLHG IPNE+TNCTE
Sbjct: 253 GISCHPGD-EQRVRSINLPYMQLGGIISPSIGKLSRLQRLALHQNSLHGTIPNELTNCTE 429
Query: 121 LRALY 125
LRALY
Sbjct: 430 LRALY 444
>TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, partial (4%)
Length = 1166
Score = 210 bits (535), Expect = 1e-54
Identities = 106/231 (45%), Positives = 149/231 (63%), Gaps = 3/231 (1%)
Frame = +1
Query: 359 ELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTE--QPLNWNDRLNI 416
E+E +G ++H +LV L GYC + R+L+Y+Y+ G+L+ LH + PL W R+NI
Sbjct: 1 EVEAIGRVRHQSLVRLLGYCAEGAHRMLVYEYVDNGNLEQWLHGDVGPCSPLTWEIRMNI 180
Query: 417 ALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVA 476
LG+A+GL YLH PK+VHRDIKSSNILL++ +SDFGLAKLL + +++TT V
Sbjct: 181 ILGTAKGLTYLHEGLEPKVVHRDIKSSNILLSKKWNAKVSDFGLAKLLGSDSSYITTRVM 360
Query: 477 GTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKE 536
GTFGY+APEY +G E+SDVYSFG+L++EL+TG+ P D S +N+V W+ +
Sbjct: 361 GTFGYVAPEYASTGMLNERSDVYSFGILIMELITGRNPVDYSRPPEEVNLVDWLKKMVSN 540
Query: 537 NRLEDVVDRRCTDADAG-TLEVILELAARCTDANADDRPSMNQVLQLLEQE 586
E V+D + + L+ L +A RCTD NA RP M V+ +LE E
Sbjct: 541 RNPEGVLDPKLPEKPTSRALKRALLVALRCTDPNAQKRPKMGHVIHMLEAE 693
>TC228048 homologue to UP|Q8LA44 (Q8LA44) Receptor protein kinase-like
protein, partial (46%)
Length = 1208
Score = 210 bits (534), Expect = 2e-54
Identities = 113/255 (44%), Positives = 171/255 (66%), Gaps = 3/255 (1%)
Frame = +2
Query: 333 VMNDCGTFAVKRI-DRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYL 391
+++D AVKR+ D + G D F+ E+E++ H NL+ L G+C P+ RLL+Y Y+
Sbjct: 5 ILSDGTLLAVKRLKDGNAIGGDIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYM 184
Query: 392 AIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENM 451
+ GS+ L + L+W R IALG+ARGL YLH +C PKI+HRD+K++NILL++
Sbjct: 185 SNGSVASRL--KGKPVLDWGTRKQIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYC 358
Query: 452 EPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTG 511
E + DFGLAKLL +D+HVTT V GT G++APEYL +G+++EK+DV+ FG+LLLEL+TG
Sbjct: 359 EAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITG 538
Query: 512 KRPTD-PSFANRGLNVVGWMNTLQKENRLEDVVDRRC-TDADAGTLEVILELAARCTDAN 569
+R + AN+ ++ W+ L +E +LE +VD+ T+ D LE I+++A CT
Sbjct: 539 QRALEFGKAANQKGAMLDWVRKLHQEKKLELLVDKDLKTNYDRIELEEIVQVALLCTQYL 718
Query: 570 ADDRPSMNQVLQLLE 584
RP M++V+++LE
Sbjct: 719 PGHRPKMSEVVRMLE 763
>TC218443 similar to GB|BAB85647.1|19032343|AB076907 inflorescence and root
apices receptor-like kinase {Arabidopsis thaliana;} ,
partial (39%)
Length = 1216
Score = 203 bits (517), Expect = 2e-52
Identities = 132/376 (35%), Positives = 209/376 (55%), Gaps = 16/376 (4%)
Frame = +2
Query: 157 GAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSF 216
GA+P L L +L NLS N GE+P G +T +S GN LCG + K C
Sbjct: 8 GALPKQLANLANLLTFNLSHNNLQGELPAGGFFNTITPSSVSGNPSLCGAAVNKSCPAVL 187
Query: 217 GFPVVI-PHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKER- 274
P+V+ P+ +D + H ++ I A+ +G ++ + + I +L+ + R
Sbjct: 188 PKPIVLNPNTSTDTGPSSLPPNLGHKRIILSISALIAIGAAAVIVIGVISITVLNLRVRS 367
Query: 275 ------AVMRYT---DVKKQVDPEAST-KLITFHGDLPYTSSEIIEKLESLEEEDIVGSG 324
A + ++ + + +A++ KL+ F G+ ++S L ++ +G G
Sbjct: 368 STPRDAAALTFSAGDEFSRSPTTDANSGKLVMFSGEPDFSSGAHA----LLNKDCELGRG 535
Query: 325 GFGTVYRMVMNDCGTFAVKRID-RSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSA 383
GFG VY+ V+ D + A+K++ S + FER+++ LG I+H NLV L GY S
Sbjct: 536 GFGAVYQTVLRDGHSVAIKKLTVSSLVKSQEDFERKVKKLGKIRHQNLVELEGYYWTTSL 715
Query: 384 RLLIYDYLAIGSLDDLLHENT-EQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKS 442
+LLIY+YL+ GSL LHE + L+WN+R N+ LG+A+ LA+LHH I+H +IKS
Sbjct: 716 QLLIYEYLSGGSLYKHLHEGSGGNFLSWNERFNVILGTAKALAHLHHS---NIIHYNIKS 886
Query: 443 SNILLNENMEPHISDFGLAKLLVDEDAHV-TTVVAGTFGYLAPEY-LQSGRATEKSDVYS 500
+N+LL+ EP + DFGLA+LL D +V ++ + GY+APE+ ++ + TEK DV
Sbjct: 887 TNVLLDSYGEPKVGDFGLARLLPMLDRYVLSSKIQSALGYMAPEFACKTVKITEKCDVLW 1066
Query: 501 FGVLLLELVTGKRPTD 516
VL+LE+VTGKRP +
Sbjct: 1067VRVLVLEIVTGKRPVE 1114
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 202 bits (513), Expect = 5e-52
Identities = 109/272 (40%), Positives = 174/272 (63%), Gaps = 4/272 (1%)
Frame = +1
Query: 321 VGSGGFGTVYRMVMNDCGTFAVKRID-RSREGCDQVFERELEILGSIKHINLVNLRGYCR 379
+G GGFG VY+ +D AVK++ +SR+G ++ F E+ ++ +++H +LV L G C
Sbjct: 13 IGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQG-NREFLNEIGMISALQHPHLVKLYGCCV 189
Query: 380 LPSARLLIYDYLAIGSLDDLLH--ENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVH 437
LL+Y+Y+ SL L E + L+W R I +G ARGLAYLH E KIVH
Sbjct: 190 EGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIVH 369
Query: 438 RDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSD 497
RDIK++N+LL++++ P ISDFGLAKL +++ H++T +AGTFGY+APEY G T+K+D
Sbjct: 370 RDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHISTRIAGTFGYMAPEYAMHGYLTDKAD 549
Query: 498 VYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRC-TDADAGTLE 556
VYSFG++ LE++ G+ T +V+ W + L+++ + D+VDRR + +
Sbjct: 550 VYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKEEAL 729
Query: 557 VILELAARCTDANADDRPSMNQVLQLLEQEVM 588
V++++A CT+ A RP+M+ V+ +LE +++
Sbjct: 730 VMIKVALLCTNVTAALRPTMSSVVSMLEGKIV 825
>TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor
(Brassinosteroid LRR receptor kinase) , partial (14%)
Length = 880
Score = 202 bits (513), Expect = 5e-52
Identities = 108/242 (44%), Positives = 151/242 (61%), Gaps = 15/242 (6%)
Frame = +3
Query: 359 ELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTE----QPLNWNDRL 414
E+E LG IKH NLV L GYC++ RLL+Y+Y+ GSL+++LH + + L W +R
Sbjct: 6 EMETLGKIKHRNLVPLLGYCKVGEERLLVYEYMEYGSLEEMLHGRIKTRDRRILTWEERK 185
Query: 415 NIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVT-T 473
IA G+A+GL +LHH C P I+HRD+KSSN+LL+ ME +SDFG+A+L+ D H++ +
Sbjct: 186 KIARGAAKGLCFLHHNCIPHIIHRDMKSSNVLLDHEMESRVSDFGMARLISALDTHLSVS 365
Query: 474 VVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTL 533
+AGT GY+ PEY QS R T K DVYSFGV++LEL++GKRPTD N+VGW
Sbjct: 366 TLAGTPGYVPPEYYQSFRCTVKGDVYSFGVVMLELLSGKRPTDKEDFG-DTNLVGWAKIK 542
Query: 534 QKENRLEDVVDRRCTDADAGTLEV----------ILELAARCTDANADDRPSMNQVLQLL 583
+E + +V+D A GT E LE+ +C D RP+M QV+ +L
Sbjct: 543 VREGKQMEVIDNDLLLATQGTDEAEAKEVKEMIRYLEITLQCVDDLPSRRPNMLQVVAML 722
Query: 584 EQ 585
+
Sbjct: 723 RE 728
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 201 bits (512), Expect = 6e-52
Identities = 121/303 (39%), Positives = 183/303 (59%), Gaps = 13/303 (4%)
Frame = +2
Query: 295 ITFHGDLPY--TSSEIIEKLESLEEEDIVGSGGFGTVYRMVMND-------CGTFAVKRI 345
I+F G Y T E+ E S +++G GGFG VY+ ++D T AVKR+
Sbjct: 5 ISFAGSKLYAFTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRL 184
Query: 346 DRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTE 405
D + + E+ LG ++H +LV L GYC RLL+Y+Y+ GSL++ L
Sbjct: 185 DLDGLQGHREWLAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYS 364
Query: 406 QPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLV 465
+ W+ R+ IALG+A+GLA+LH P +++RD K+SNILL+ + +SDFGLAK
Sbjct: 365 AAMPWSTRMKIALGAAKGLAFLHEADKP-VIYRDFKASNILLDSDFTAKLSDFGLAKDGP 541
Query: 466 D-EDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGL 524
+ ED HVTT + GT GY APEY+ +G T KSDVYS+GV+LLEL+TG+R D S +N G
Sbjct: 542 EGEDTHVTTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGK 721
Query: 525 NVVGWMN-TLQKENRLEDVVDRRCTD--ADAGTLEVILELAARCTDANADDRPSMNQVLQ 581
++V W L+ + ++ +++DRR G ++V + LA +C + + RP+M+ V++
Sbjct: 722 SLVEWARPLLRDQKKVYNIIDRRLEGQFPMKGAMKVAM-LAFKCLSHHPNARPTMSDVIK 898
Query: 582 LLE 584
+LE
Sbjct: 899 VLE 907
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,694,728
Number of Sequences: 63676
Number of extensions: 357442
Number of successful extensions: 4084
Number of sequences better than 10.0: 1123
Number of HSP's better than 10.0 without gapping: 2981
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3218
length of query: 603
length of database: 12,639,632
effective HSP length: 103
effective length of query: 500
effective length of database: 6,081,004
effective search space: 3040502000
effective search space used: 3040502000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0170.5


